"cugbp, Elav-like family member 3a"
ZFIN 
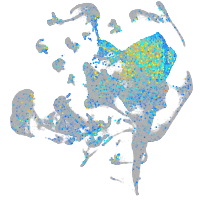
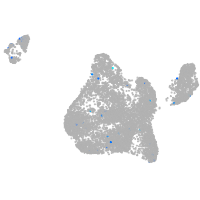
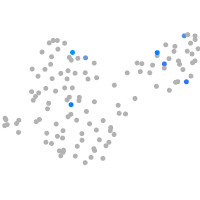
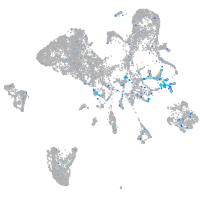
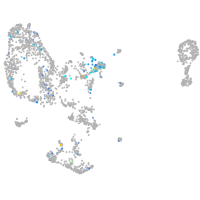
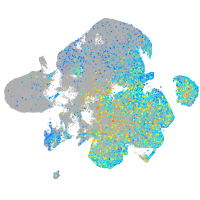
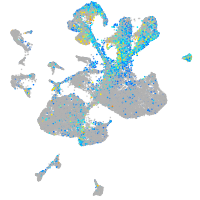
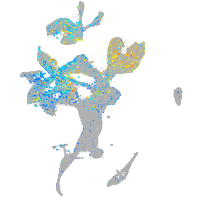
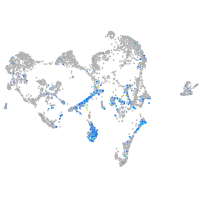
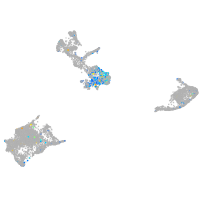
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod1 | 0.373 | aldob | -0.142 |
| insm1a | 0.358 | gapdh | -0.138 |
| myt1b | 0.329 | gamt | -0.132 |
| scg3 | 0.328 | ahcy | -0.121 |
| pax6b | 0.325 | fbp1b | -0.120 |
| mir7a-1 | 0.298 | gatm | -0.120 |
| id4 | 0.292 | aldh7a1 | -0.114 |
| nkx2.2a | 0.286 | nupr1b | -0.114 |
| syt1a | 0.277 | mat1a | -0.112 |
| vamp2 | 0.274 | apoa4b.1 | -0.110 |
| gnao1a | 0.273 | aldh6a1 | -0.109 |
| si:dkey-153k10.9 | 0.268 | cx32.3 | -0.108 |
| pcsk1 | 0.265 | hdlbpa | -0.106 |
| rnasekb | 0.264 | gstt1a | -0.106 |
| LOC100537384 | 0.263 | bhmt | -0.106 |
| scgn | 0.257 | scp2a | -0.106 |
| pcsk1nl | 0.255 | eno3 | -0.106 |
| lysmd2 | 0.255 | apoa1b | -0.101 |
| gpc1a | 0.255 | agxtb | -0.100 |
| tox | 0.255 | mgst1.2 | -0.099 |
| jagn1a | 0.254 | apoc2 | -0.099 |
| isl1 | 0.252 | suclg2 | -0.098 |
| tmsb | 0.252 | aqp12 | -0.098 |
| insm1b | 0.249 | ckba | -0.097 |
| kcnj11 | 0.245 | abat | -0.097 |
| dlb | 0.244 | gstr | -0.097 |
| dpysl2b | 0.242 | suclg1 | -0.097 |
| vat1 | 0.240 | glud1b | -0.096 |
| rprmb | 0.233 | gnmt | -0.096 |
| atp6v0cb | 0.233 | agxta | -0.096 |
| tspan7b | 0.232 | cebpd | -0.096 |
| gdi1 | 0.230 | gpx4a | -0.096 |
| pcsk2 | 0.230 | rgn | -0.095 |
| pcdh20 | 0.229 | gstp1 | -0.095 |
| rasd4 | 0.228 | ttc36 | -0.093 |