cyclin-dependent kinase 8
ZFIN 
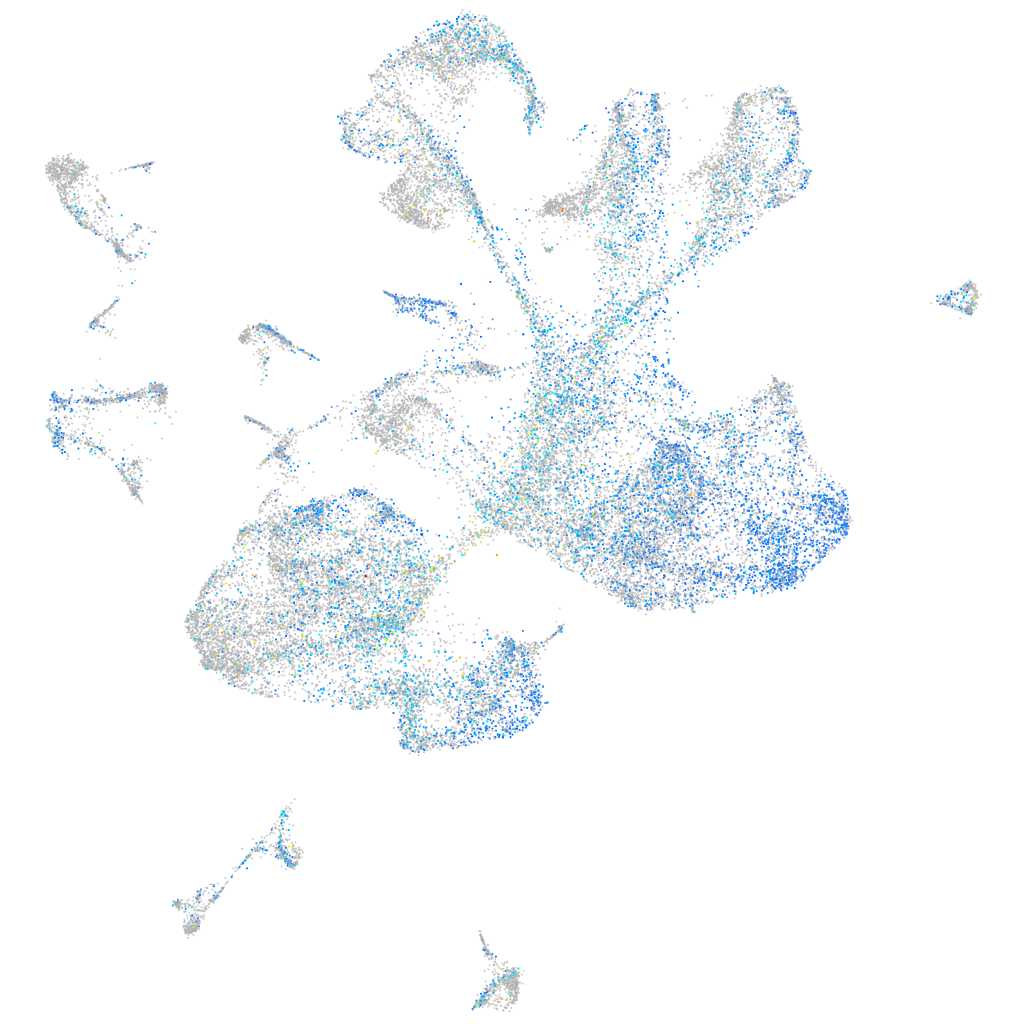
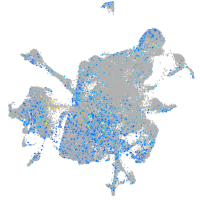
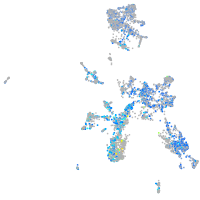
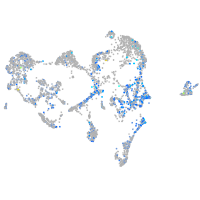
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpabb | 0.172 | CR383676.1 | -0.098 |
| hnrnpa0l | 0.160 | ppdpfb | -0.097 |
| hnrnpaba | 0.158 | apoa2 | -0.078 |
| khdrbs1a | 0.157 | cx43 | -0.077 |
| ddx39ab | 0.148 | glula | -0.077 |
| snrpd2 | 0.146 | mt2 | -0.073 |
| hnrnpa0b | 0.146 | nupr1a | -0.073 |
| snrpg | 0.146 | atp1a1b | -0.072 |
| cirbpa | 0.146 | pvalb1 | -0.072 |
| eif4a1a | 0.144 | cox4i2 | -0.071 |
| abcf1 | 0.144 | zgc:153704 | -0.071 |
| hmga1a | 0.140 | apoa1b | -0.070 |
| snrpd1 | 0.140 | nanos1 | -0.070 |
| alyref | 0.139 | slc4a4a | -0.069 |
| si:ch211-222l21.1 | 0.139 | cebpd | -0.068 |
| apex1 | 0.138 | ptn | -0.067 |
| seta | 0.137 | si:ch211-66e2.5 | -0.066 |
| snrpf | 0.137 | acbd7 | -0.066 |
| psmb7 | 0.137 | dap1b | -0.066 |
| ilf2 | 0.137 | znf704 | -0.066 |
| top1l | 0.137 | sesn1 | -0.066 |
| cct5 | 0.134 | prss59.2 | -0.066 |
| snrpe | 0.134 | IGLON5 | -0.066 |
| syncrip | 0.132 | gapdhs | -0.065 |
| hdac1 | 0.132 | slc1a2b | -0.065 |
| ybx1 | 0.131 | mdkb | -0.064 |
| cct3 | 0.131 | slc3a2a | -0.064 |
| setb | 0.131 | zgc:92606 | -0.064 |
| psma3 | 0.131 | cldn7a | -0.063 |
| ran | 0.131 | pvalb2 | -0.063 |
| hnrnph1l | 0.130 | actc1b | -0.062 |
| hmgb2b | 0.130 | efhd1 | -0.062 |
| nono | 0.129 | lpin1 | -0.062 |
| snrpb | 0.128 | FQ323156.1 | -0.061 |
| tubb2b | 0.127 | aldocb | -0.061 |