cyclin-dependent kinase 15
ZFIN 
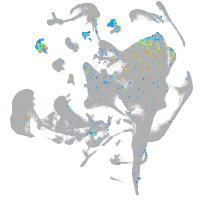
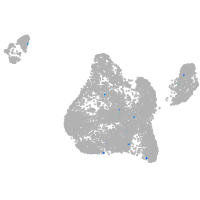
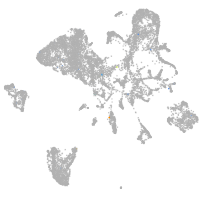
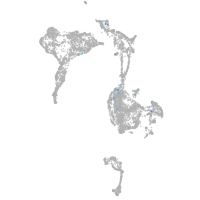
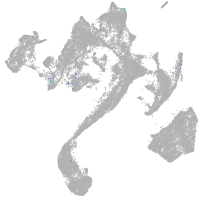
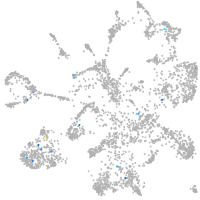
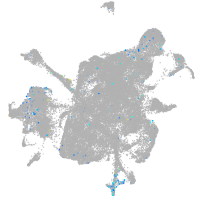
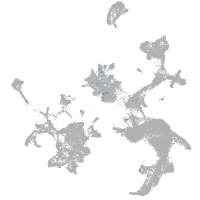
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmela | 0.296 | CABZ01021592.1 | -0.133 |
| dct | 0.291 | si:ch211-222l21.1 | -0.128 |
| tyrp1b | 0.288 | ptmaa | -0.116 |
| tyrp1a | 0.284 | si:ch73-1a9.3 | -0.115 |
| slc24a5 | 0.280 | tuba1c | -0.112 |
| zgc:91968 | 0.280 | paics | -0.111 |
| SPAG9 | 0.278 | si:dkey-251i10.2 | -0.111 |
| si:ch73-389b16.1 | 0.274 | hmgb1b | -0.107 |
| slc3a2a | 0.271 | gpm6aa | -0.107 |
| slc45a2 | 0.271 | tmsb | -0.105 |
| tyr | 0.270 | si:ch73-281n10.2 | -0.105 |
| oca2 | 0.268 | hmgn2 | -0.105 |
| tspan36 | 0.268 | mdh1aa | -0.103 |
| mtbl | 0.268 | hbbe1.1 | -0.102 |
| kita | 0.267 | hbbe1.3 | -0.101 |
| aadac | 0.259 | stmn1a | -0.100 |
| tfap2e | 0.256 | hbae3 | -0.099 |
| rabl6b | 0.254 | si:ch211-288g17.3 | -0.097 |
| prkar1b | 0.252 | nova2 | -0.096 |
| kcnj13 | 0.247 | hmgb2b | -0.093 |
| lamp1a | 0.247 | stmn1b | -0.092 |
| mlpha | 0.245 | ptmab | -0.091 |
| bace2 | 0.243 | hmgb3a | -0.090 |
| slc37a2 | 0.243 | pbx3b | -0.089 |
| atp6v0ca | 0.242 | epb41a | -0.089 |
| rab38 | 0.241 | pvalb1 | -0.089 |
| tspan10 | 0.239 | gng2 | -0.088 |
| mitfa | 0.235 | sox11b | -0.088 |
| gstt1a | 0.234 | tmeff1b | -0.085 |
| si:ch211-195b13.1 | 0.232 | elavl3 | -0.085 |
| opn5 | 0.229 | uraha | -0.085 |
| slc7a5 | 0.228 | zc4h2 | -0.084 |
| si:zfos-943e10.1 | 0.227 | gng3 | -0.084 |
| pah | 0.224 | h3f3a | -0.083 |
| pcdh10a | 0.222 | zgc:101840 | -0.082 |