cell division cycle 6 homolog (S. cerevisiae)
ZFIN 
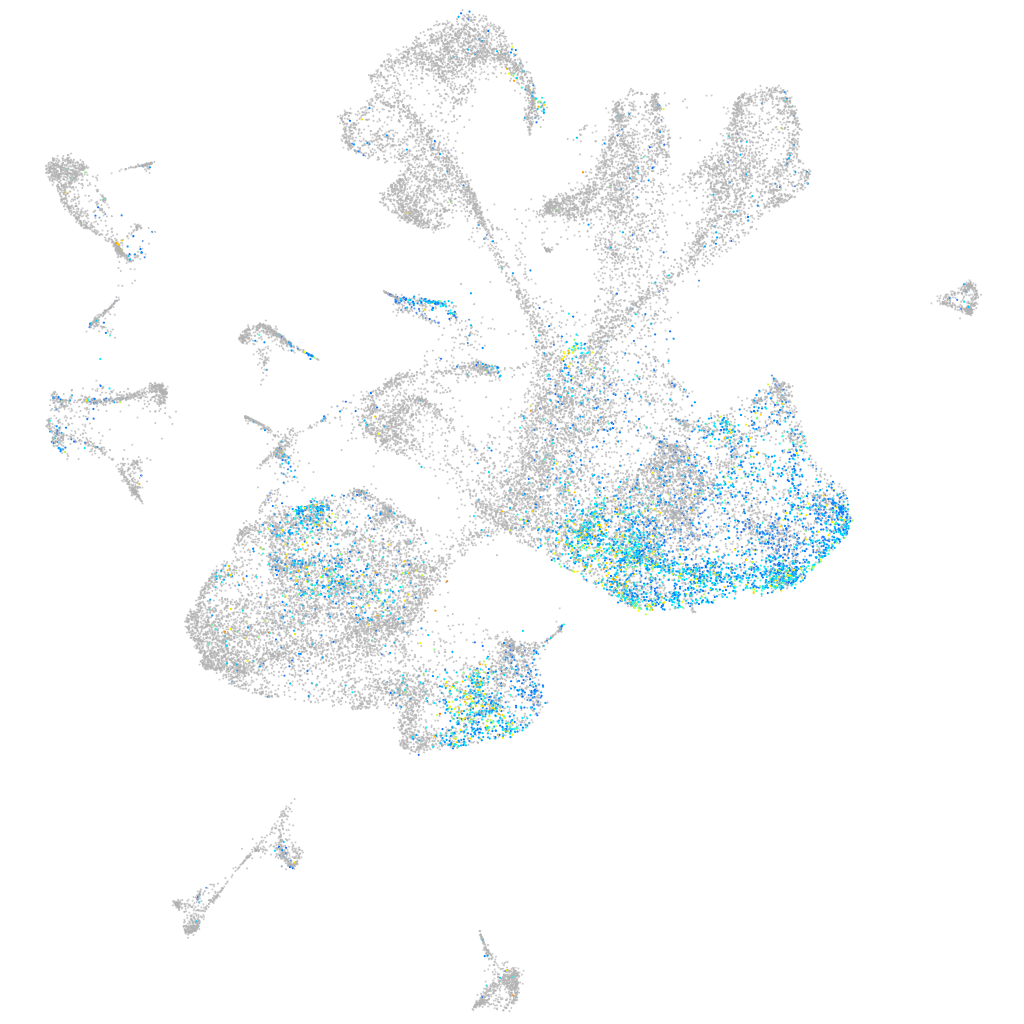
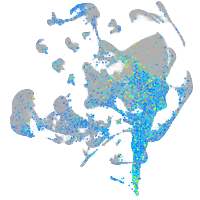
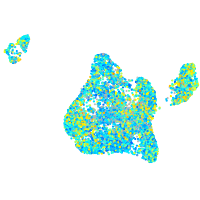
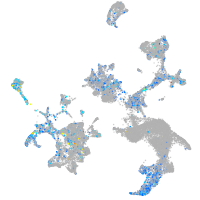
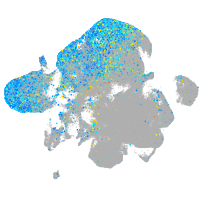
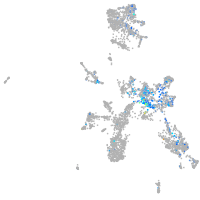
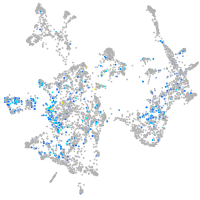
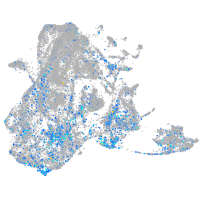
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slbp | 0.568 | ckbb | -0.246 |
| pcna | 0.554 | rtn1a | -0.207 |
| mcm5 | 0.529 | gpm6aa | -0.204 |
| fen1 | 0.521 | gpm6ab | -0.195 |
| cdca7a | 0.520 | rnasekb | -0.193 |
| nasp | 0.515 | elavl3 | -0.193 |
| zgc:110540 | 0.497 | myt1b | -0.185 |
| hells | 0.495 | si:dkeyp-75h12.5 | -0.183 |
| mcm4 | 0.495 | h2afx1 | -0.180 |
| mcm2 | 0.494 | CR383676.1 | -0.177 |
| mcm6 | 0.487 | stmn1b | -0.175 |
| chaf1a | 0.474 | ppdpfb | -0.173 |
| rpa2 | 0.473 | pvalb1 | -0.170 |
| mcm3 | 0.471 | gapdhs | -0.170 |
| rpa3 | 0.464 | gng3 | -0.170 |
| lig1 | 0.464 | pvalb2 | -0.166 |
| mcm7 | 0.462 | elavl4 | -0.166 |
| dut | 0.454 | calm1a | -0.165 |
| CABZ01005379.1 | 0.454 | h1f0 | -0.161 |
| gins2 | 0.449 | atp6v0cb | -0.161 |
| dnajc9 | 0.442 | sncb | -0.160 |
| rrm2 | 0.441 | marcksl1b | -0.160 |
| stmn1a | 0.440 | atp6v1e1b | -0.158 |
| ccne2 | 0.437 | cotl1 | -0.155 |
| esco2 | 0.435 | slc1a2b | -0.154 |
| gmnn | 0.434 | vamp2 | -0.154 |
| rrm1 | 0.431 | fez1 | -0.154 |
| e2f7 | 0.421 | cadm4 | -0.154 |
| msh6 | 0.421 | dpysl3 | -0.152 |
| selenoh | 0.417 | stxbp1a | -0.151 |
| asf1ba | 0.414 | CU467822.1 | -0.150 |
| unga | 0.408 | COX3 | -0.150 |
| dhfr | 0.405 | dpysl2b | -0.150 |
| rfc2 | 0.402 | tmsb | -0.149 |
| cdca7b | 0.400 | gnao1a | -0.148 |