CDC42 effector protein (Rho GTPase binding) 4b
ZFIN 
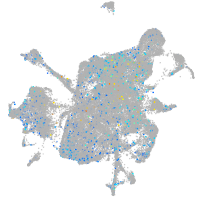
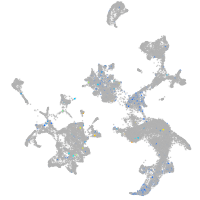
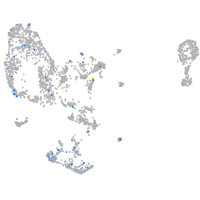
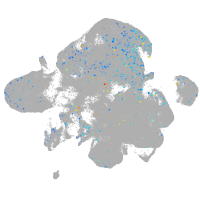
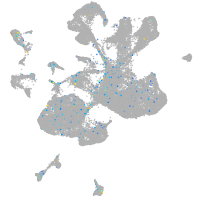
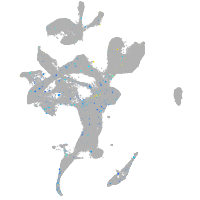
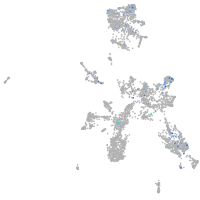
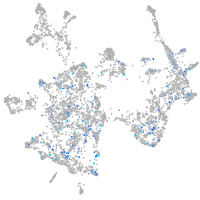
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:195212 | 0.171 | tmsb4x | -0.082 |
| slc47a3 | 0.160 | acta2 | -0.079 |
| tnni1a | 0.160 | tpm1 | -0.074 |
| or112-1 | 0.150 | myl9a | -0.073 |
| pcdh2g6 | 0.150 | csrp1b | -0.069 |
| or137-3 | 0.142 | ckba | -0.065 |
| crygm2d20 | 0.142 | igfbp7 | -0.064 |
| zgc:174314 | 0.133 | myh11a | -0.062 |
| CR354444.1 | 0.129 | tagln | -0.060 |
| AL929171.1 | 0.122 | tgfb3 | -0.060 |
| XLOC-014488 | 0.122 | gucy1a1 | -0.060 |
| zgc:171497 | 0.121 | lmod1b | -0.059 |
| upp2 | 0.121 | myl6 | -0.058 |
| proser2 | 0.118 | tpm2 | -0.057 |
| mmp16a | 0.118 | fhl2a | -0.057 |
| XLOC-011687 | 0.117 | cnn1b | -0.055 |
| kcnj12a | 0.116 | pkma | -0.055 |
| LOC110439394 | 0.115 | cald1b | -0.054 |
| si:ch73-252i11.1 | 0.114 | acta1a | -0.053 |
| prcc | 0.114 | ldha | -0.051 |
| LOC103908899 | 0.110 | dusp2 | -0.050 |
| tom1l2a | 0.110 | pgm5 | -0.050 |
| vwf | 0.110 | tuba8l2 | -0.050 |
| arhgef28 | 0.109 | fbxl22 | -0.049 |
| synrg | 0.108 | mcamb | -0.049 |
| CR936975.1 | 0.108 | cavin2b | -0.049 |
| cyp11c1 | 0.108 | fxyd1 | -0.048 |
| poc1bl | 0.107 | eno3 | -0.048 |
| psmc3ip | 0.106 | gpc6a | -0.048 |
| CABZ01088229.1 | 0.105 | chac1 | -0.048 |
| BX088535.1 | 0.104 | cxcl14 | -0.048 |
| cd74a | 0.104 | gapdhs | -0.047 |
| zgc:171711 | 0.104 | mef2aa | -0.047 |
| rln | 0.103 | rbpms2b | -0.047 |
| CU655961.4 | 0.102 | eno1a | -0.046 |