coiled-coil domain containing 88B
ZFIN 
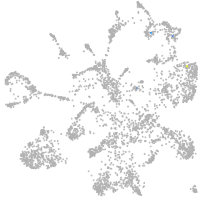
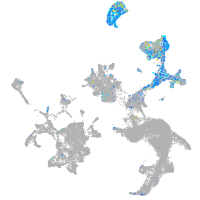
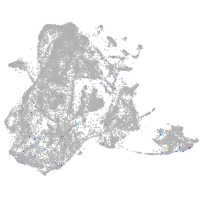
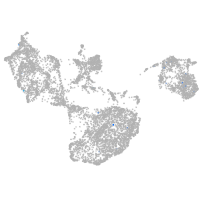
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL935300.2 | 0.121 | rtn1a | -0.016 |
| hhex | 0.060 | tuba1c | -0.016 |
| XLOC-031233 | 0.057 | elavl3 | -0.015 |
| RF00407 | 0.056 | ptmaa | -0.015 |
| XLOC-019047 | 0.050 | gpm6aa | -0.014 |
| XLOC-022251 | 0.048 | gpm6ab | -0.014 |
| RF00019 | 0.047 | hmgb3a | -0.014 |
| dysf | 0.044 | tubb2b | -0.013 |
| FO681472.1 | 0.044 | nova2 | -0.013 |
| LOC110437769 | 0.043 | stmn1b | -0.013 |
| cdh15 | 0.042 | zfhx3 | -0.013 |
| cdaa | 0.042 | atp6v0cb | -0.013 |
| abi3bpa | 0.040 | actc1b | -0.012 |
| BX511028.1 | 0.039 | h3f3c | -0.012 |
| rin3 | 0.039 | pvalb2 | -0.012 |
| ighv1-4 | 0.039 | rnasekb | -0.012 |
| si:ch211-214p16.1 | 0.038 | fabp3 | -0.012 |
| smad4a | 0.036 | marcksl1a | -0.012 |
| LOC110438254 | 0.036 | tubb5 | -0.012 |
| XLOC-007706 | 0.036 | rtn1b | -0.012 |
| btr09 | 0.035 | ckbb | -0.012 |
| upp2 | 0.035 | vat1 | -0.012 |
| CABZ01109481.1 | 0.035 | tmsb2 | -0.011 |
| ssuh2.2 | 0.034 | CU467822.1 | -0.011 |
| nkx2.3 | 0.034 | gapdhs | -0.011 |
| si:dkeyp-4c4.2 | 0.033 | meis2b | -0.011 |
| LO018557.1 | 0.032 | nova1 | -0.011 |
| si:dkey-11p23.7 | 0.032 | myt1b | -0.011 |
| drl | 0.032 | CR383676.1 | -0.011 |
| slc9a3.2 | 0.031 | tnrc6c2 | -0.011 |
| XLOC-029465 | 0.031 | gng3 | -0.011 |
| CABZ01086127.1 | 0.030 | si:ch1073-429i10.3.1 | -0.010 |
| she | 0.030 | CU634008.1 | -0.010 |
| dmrt1 | 0.029 | dpysl3 | -0.010 |
| parm1 | 0.029 | sncb | -0.010 |