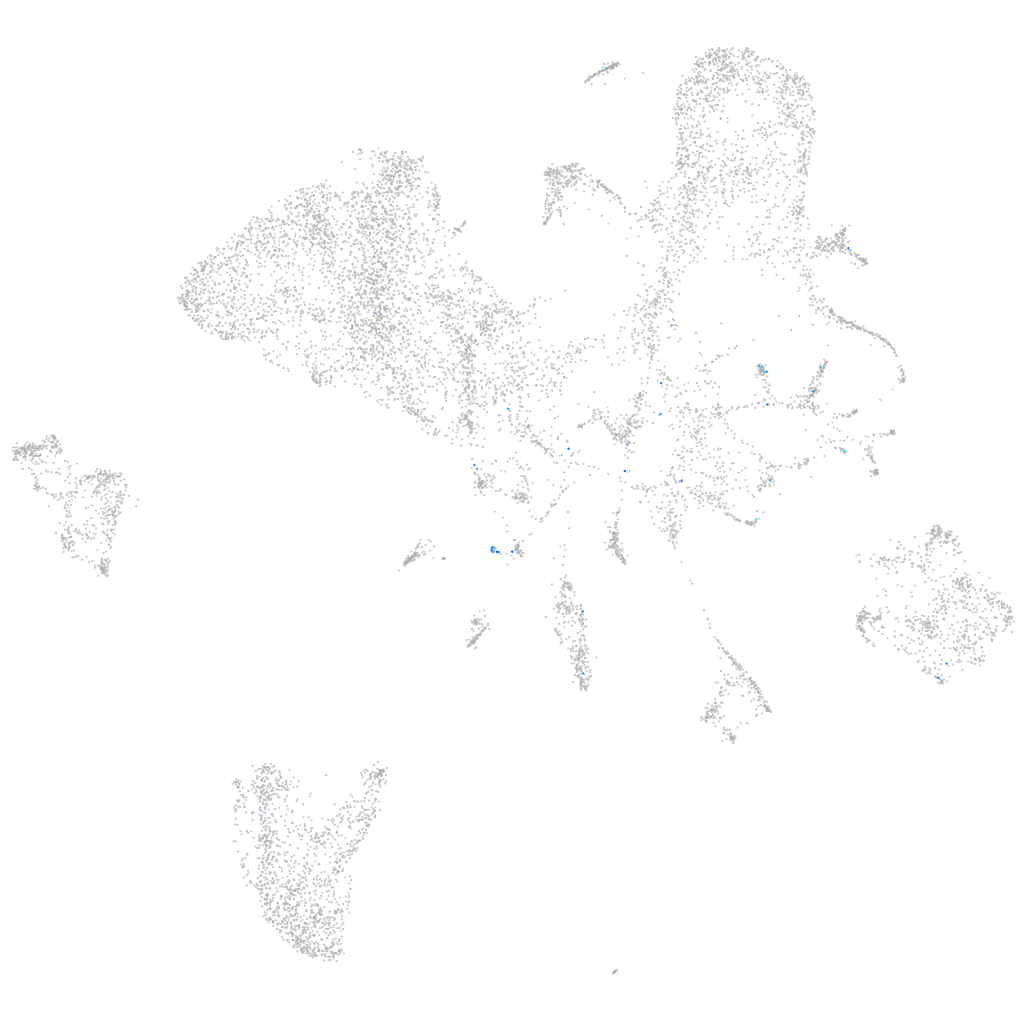
coiled-coil domain containing 65
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| spata18 | 0.315 | ahcy | -0.050 |
| LOC103911624 | 0.302 | gamt | -0.047 |
| XLOC-039765 | 0.300 | gatm | -0.042 |
| zgc:195356 | 0.295 | gapdh | -0.042 |
| cfap126 | 0.275 | dap | -0.041 |
| tekt1 | 0.273 | bhmt | -0.041 |
| gmnc | 0.273 | nupr1b | -0.040 |
| cdc25d | 0.266 | mat1a | -0.040 |
| LOC101884915 | 0.265 | apoa4b.1 | -0.038 |
| CFAP77 | 0.259 | apoa1b | -0.038 |
| si:dkey-76p14.2 | 0.257 | fabp3 | -0.037 |
| spaca9 | 0.254 | fbp1b | -0.037 |
| si:dkey-148f10.4 | 0.250 | cbr1l | -0.037 |
| tex9 | 0.244 | apoc2 | -0.037 |
| zc2hc1c | 0.240 | scp2a | -0.037 |
| tex36 | 0.234 | gstt1a | -0.036 |
| dydc2 | 0.233 | glud1b | -0.036 |
| ccdc181 | 0.223 | afp4 | -0.036 |
| capsla | 0.221 | cdo1 | -0.036 |
| daw1 | 0.216 | cx32.3 | -0.035 |
| nek10 | 0.214 | agxtb | -0.035 |
| tbata | 0.214 | rdh1 | -0.035 |
| pacrg | 0.212 | tdo2a | -0.035 |
| dnah5l | 0.212 | mgst1.2 | -0.035 |
| ccdc146 | 0.211 | slc25a34 | -0.034 |
| dnajb13 | 0.211 | acmsd | -0.034 |
| ccdc151 | 0.208 | gnmt | -0.034 |
| ribc2 | 0.200 | aldh7a1 | -0.034 |
| si:ch211-155m12.5 | 0.199 | adka | -0.034 |
| efhc2 | 0.199 | nipsnap3a | -0.033 |
| ccdc173 | 0.197 | cyp8b1 | -0.033 |
| enkur | 0.195 | hspe1 | -0.032 |
| cfap100 | 0.194 | slc16a10 | -0.032 |
| cfap45 | 0.193 | hdlbpa | -0.032 |
| spag1b | 0.192 | rgn | -0.032 |