coiled-coil domain containing 22
ZFIN 
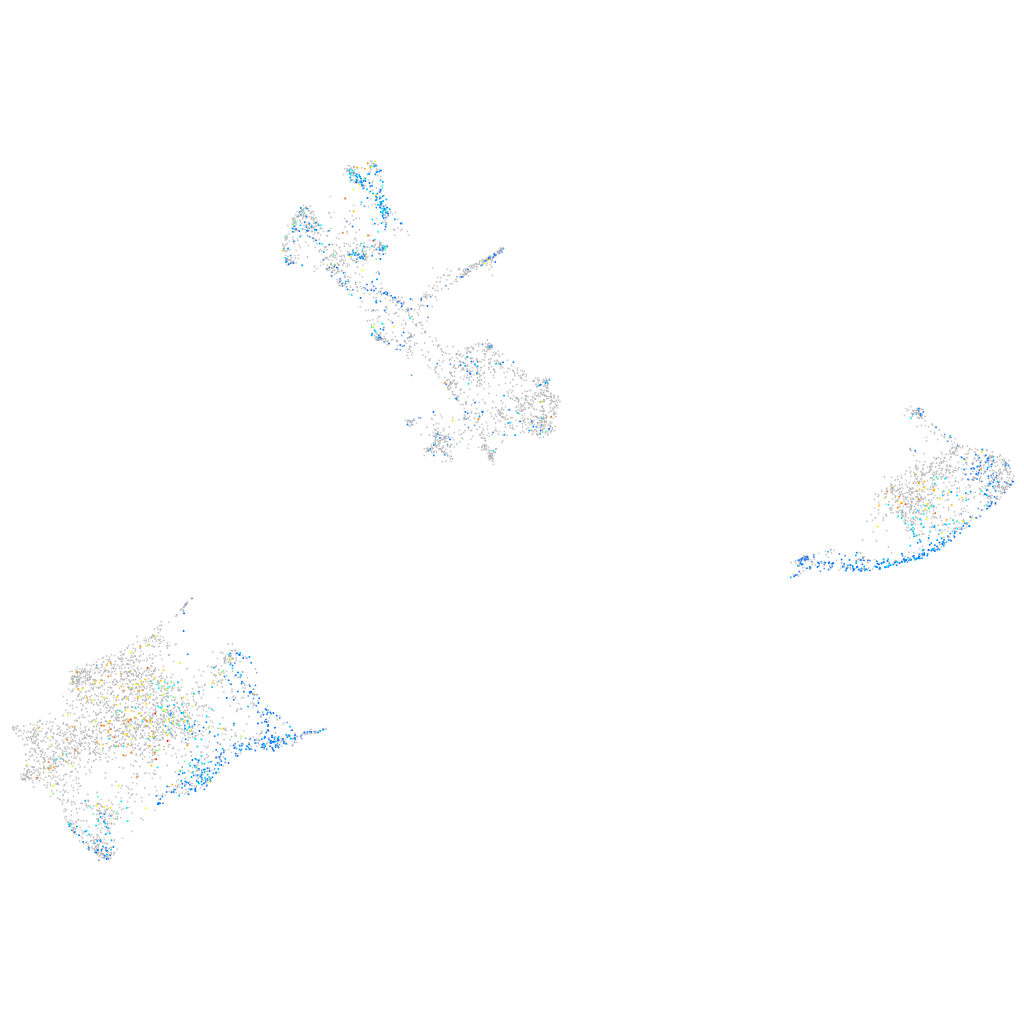
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rab32a | 0.160 | ptmaa | -0.108 |
| zgc:110239 | 0.150 | gpm6aa | -0.092 |
| nme4 | 0.145 | pvalb1 | -0.092 |
| agtrap | 0.143 | si:ch211-222l21.1 | -0.092 |
| BX571715.1 | 0.140 | tuba1c | -0.091 |
| pnp4a | 0.138 | si:ch73-1a9.3 | -0.084 |
| smim29 | 0.138 | marcksl1a | -0.084 |
| glulb | 0.137 | actc1b | -0.082 |
| impdh1b | 0.135 | nova2 | -0.082 |
| rab38 | 0.133 | stmn1b | -0.076 |
| naga | 0.133 | pvalb2 | -0.075 |
| psph | 0.132 | zc4h2 | -0.073 |
| canx | 0.131 | hmgb3a | -0.073 |
| syngr1a | 0.131 | stmn1a | -0.072 |
| dhrs12 | 0.131 | mylz3 | -0.071 |
| sypl2b | 0.130 | hmgb1b | -0.070 |
| gdi2 | 0.130 | icn | -0.069 |
| ctsba | 0.130 | hbae3 | -0.068 |
| trpm1b | 0.130 | elavl3 | -0.068 |
| atp11a | 0.129 | hbbe1.3 | -0.066 |
| eno3 | 0.129 | epb41a | -0.066 |
| bloc1s4 | 0.129 | tmsb | -0.065 |
| sptlc2a | 0.128 | gng2 | -0.064 |
| pah | 0.128 | ptmab | -0.064 |
| calr3b | 0.127 | mylpfa | -0.063 |
| atp6v1c1b | 0.127 | ckma | -0.062 |
| ldhba | 0.126 | foxp4 | -0.062 |
| rnaseka | 0.126 | si:ch73-281n10.2 | -0.062 |
| atp6v1ab | 0.126 | tubb5 | -0.062 |
| aldh16a1 | 0.126 | cdh2 | -0.062 |
| CABZ01032488.1 | 0.125 | ndrg2 | -0.062 |
| si:ch211-243a20.3 | 0.125 | rnasekb | -0.062 |
| rgs4 | 0.125 | neurod4 | -0.061 |
| snx2 | 0.125 | hbae1.1 | -0.061 |
| qdpra | 0.125 | s100a10b | -0.061 |