COBW domain containing
ZFIN 
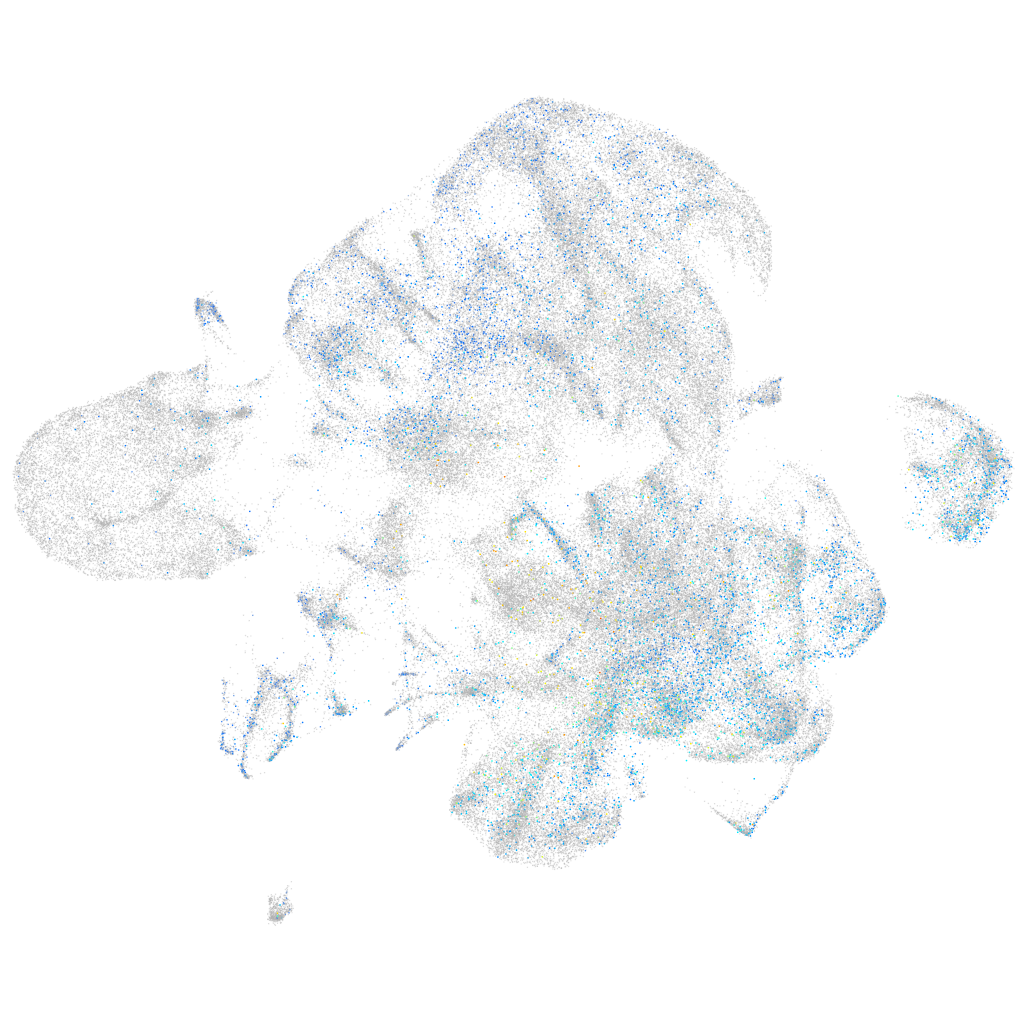
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.117 | hmgb2a | -0.076 |
| ywhag2 | 0.115 | hmga1a | -0.074 |
| stmn2a | 0.111 | cx43.4 | -0.059 |
| zgc:65894 | 0.110 | hmgb2b | -0.056 |
| gng3 | 0.108 | hspb1 | -0.056 |
| atp6v1e1b | 0.108 | NC-002333.4 | -0.054 |
| sncb | 0.107 | si:ch211-152c2.3 | -0.053 |
| mllt11 | 0.104 | id1 | -0.052 |
| tpi1b | 0.103 | zgc:110425 | -0.051 |
| tmsb2 | 0.102 | apoeb | -0.051 |
| atp6v1g1 | 0.101 | akap12b | -0.050 |
| snap25a | 0.101 | stm | -0.049 |
| calm2a | 0.100 | rps12 | -0.049 |
| calm1b | 0.097 | hnrnpa1b | -0.048 |
| syn2a | 0.096 | si:dkey-151g10.6 | -0.048 |
| calm1a | 0.096 | si:ch211-222l21.1 | -0.048 |
| gap43 | 0.096 | chaf1a | -0.047 |
| calm3a | 0.095 | sox19a | -0.046 |
| atp6v0cb | 0.095 | aldob | -0.046 |
| cdk5r2a | 0.094 | msna | -0.045 |
| calm3b | 0.093 | lig1 | -0.045 |
| rnasekb | 0.093 | rps18 | -0.045 |
| ldhba | 0.093 | CABZ01037298.1 | -0.045 |
| ppiab | 0.092 | rplp2l | -0.044 |
| atp5mc1 | 0.091 | nr6a1a | -0.044 |
| eno1a | 0.091 | ccnd1 | -0.044 |
| atp6v1h | 0.091 | XLOC-003689 | -0.044 |
| vamp2 | 0.090 | dla | -0.044 |
| atpv0e2 | 0.090 | mki67 | -0.043 |
| uchl1 | 0.090 | si:ch73-281n10.2 | -0.043 |
| tuba1c | 0.090 | rps28 | -0.043 |
| zgc:153426 | 0.089 | her15.1 | -0.043 |
| rab6bb | 0.089 | arf1 | -0.043 |
| map1aa | 0.088 | chd7 | -0.043 |
| cnrip1a | 0.087 | notch1a | -0.042 |