CACN subunit beta associated regulatory protein b
ZFIN 
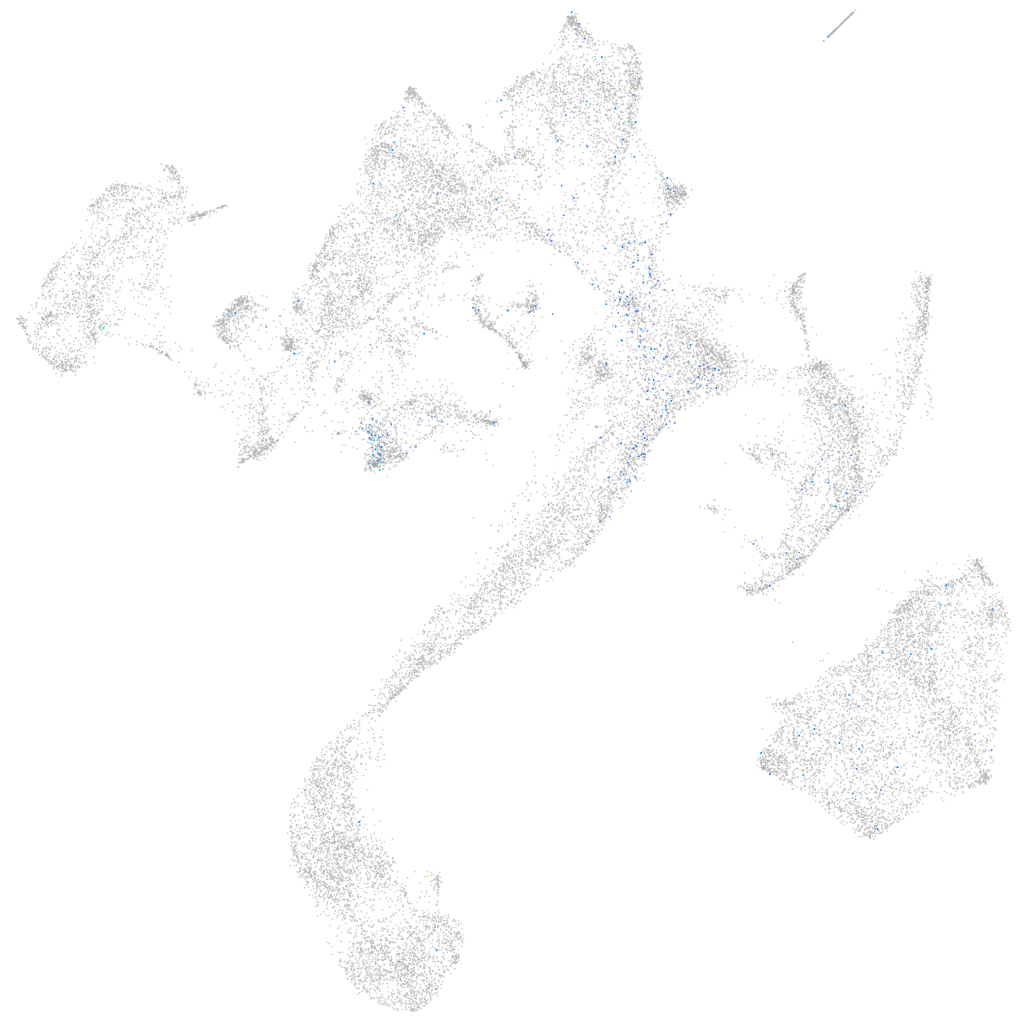
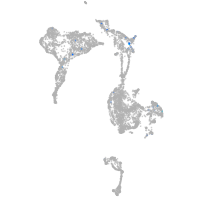
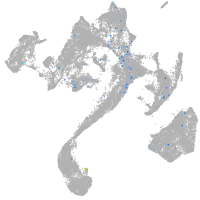
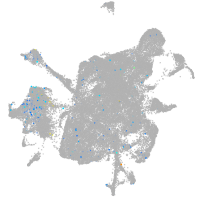
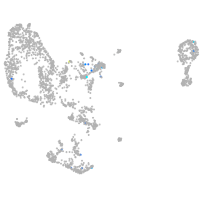
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01084564.1 | 0.218 | aldoab | -0.046 |
| taar20w | 0.185 | gapdh | -0.044 |
| BX927275.2 | 0.170 | tmem38a | -0.044 |
| gabra3 | 0.156 | ckma | -0.044 |
| synpr | 0.156 | ldb3a | -0.043 |
| zgc:101731 | 0.151 | ckmb | -0.043 |
| nwd2 | 0.148 | neb | -0.042 |
| gng8 | 0.146 | actc1b | -0.041 |
| mcf2a | 0.120 | atp2a1 | -0.041 |
| tacr2 | 0.116 | myom1a | -0.041 |
| nptx2a | 0.115 | ldb3b | -0.041 |
| rims3 | 0.108 | tpma | -0.040 |
| XLOC-027918 | 0.107 | tnnc2 | -0.040 |
| arhgef9a | 0.106 | srl | -0.040 |
| CABZ01041604.1 | 0.106 | acta1b | -0.040 |
| pcsk2 | 0.105 | cav3 | -0.040 |
| cadps2 | 0.104 | mylpfa | -0.040 |
| XLOC-019062 | 0.104 | desma | -0.040 |
| LOC103910472 | 0.103 | tnnt3a | -0.040 |
| LOC108181225 | 0.103 | eno3 | -0.040 |
| grm3 | 0.101 | casq2 | -0.039 |
| es1 | 0.100 | CABZ01078594.1 | -0.039 |
| ccl38a.4 | 0.099 | acta1a | -0.038 |
| gabbr1a | 0.099 | mybphb | -0.038 |
| cacna2d2b | 0.098 | tmem182a | -0.038 |
| nlgn3a | 0.098 | ak1 | -0.038 |
| cep170ab | 0.098 | XLOC-006515 | -0.037 |
| pik3r6a | 0.097 | mef2ca | -0.037 |
| scg2b | 0.096 | actn3a | -0.037 |
| syt1a | 0.096 | actn3b | -0.037 |
| adcyap1r1a | 0.096 | cavin4b | -0.037 |
| olfm1b | 0.095 | pabpc4 | -0.037 |
| sypa | 0.095 | CR376766.2 | -0.037 |
| ngb | 0.095 | CABZ01072309.1 | -0.036 |
| stx1b | 0.093 | smyd1a | -0.036 |