capping protein regulator and myosin 1 linker 3
ZFIN 
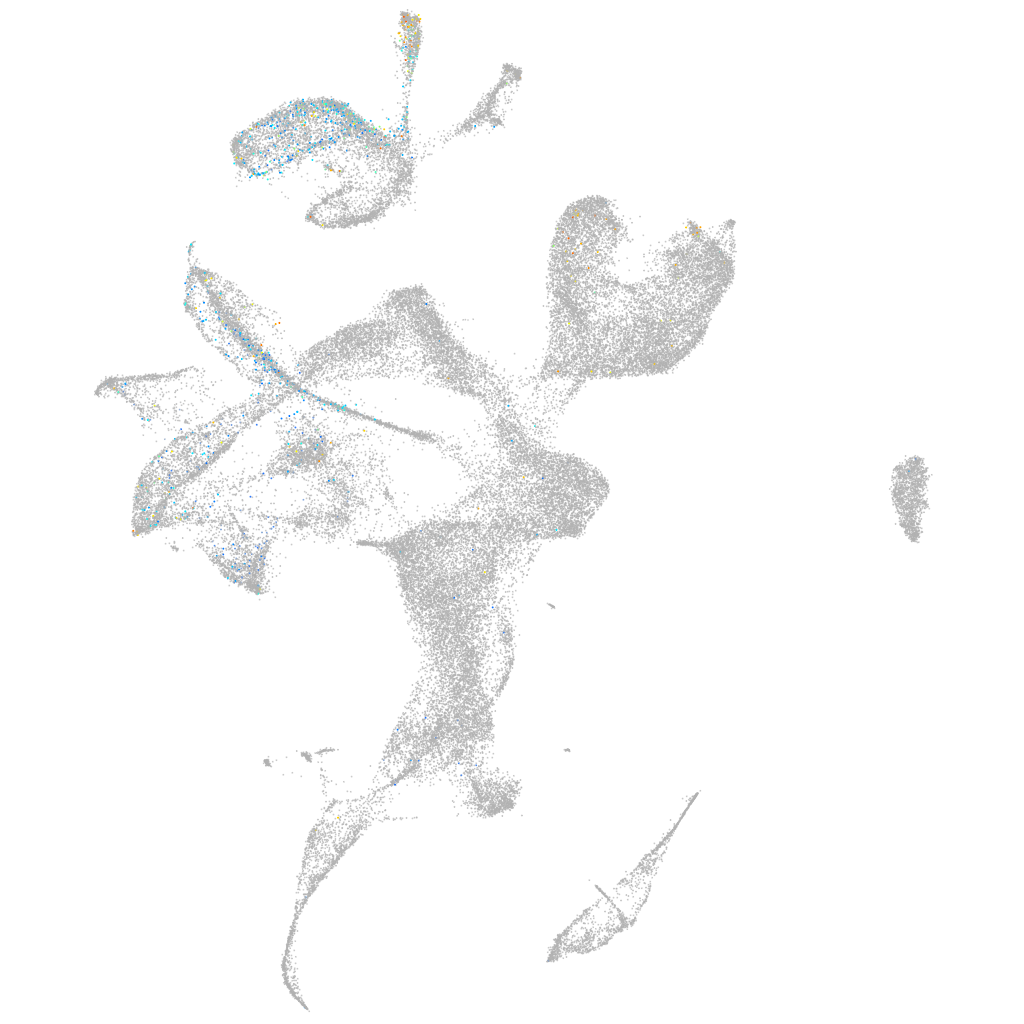
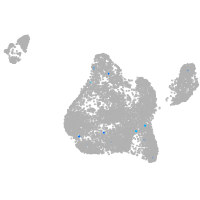
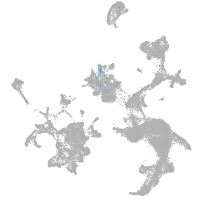
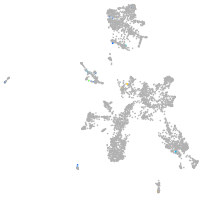
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pax10 | 0.135 | hmgb2a | -0.118 |
| elavl3 | 0.133 | pcna | -0.067 |
| slc18a3a | 0.127 | stmn1a | -0.066 |
| stxbp1a | 0.127 | mdka | -0.064 |
| stmn1b | 0.124 | hmgb2b | -0.063 |
| pvalb6 | 0.122 | rrm1 | -0.063 |
| syt1a | 0.120 | dut | -0.062 |
| tfap2a | 0.120 | ccnd1 | -0.062 |
| slc32a1 | 0.113 | tuba8l4 | -0.062 |
| meis2b | 0.113 | lbr | -0.062 |
| stx1b | 0.112 | mki67 | -0.060 |
| tfap2b | 0.111 | hmga1a | -0.060 |
| zfhx3 | 0.110 | msna | -0.060 |
| CR855337.1 | 0.110 | mcm7 | -0.059 |
| pbx1a | 0.107 | selenoh | -0.059 |
| gnao1a | 0.103 | tuba8l | -0.059 |
| cplx2 | 0.102 | ccna2 | -0.059 |
| gad1b | 0.102 | nutf2l | -0.058 |
| zgc:101840 | 0.101 | chaf1a | -0.058 |
| rbfox2 | 0.100 | ran | -0.058 |
| gldn | 0.099 | banf1 | -0.056 |
| si:ch211-129p13.1 | 0.099 | dek | -0.056 |
| tiam1a | 0.098 | cks1b | -0.055 |
| slc6a1a | 0.097 | her15.1 | -0.055 |
| sncb | 0.097 | rpa3 | -0.054 |
| nrxn1a | 0.096 | fen1 | -0.054 |
| gng3 | 0.095 | si:ch211-156b7.4 | -0.054 |
| dtnbp1b | 0.095 | ahcy | -0.054 |
| ywhag2 | 0.095 | rps20 | -0.053 |
| barhl2 | 0.094 | fabp7a | -0.053 |
| gpr158a | 0.093 | nasp | -0.053 |
| mir181b-3 | 0.093 | COX7A2 (1 of many) | -0.052 |
| sv2a | 0.092 | rpsa | -0.052 |
| pcdh19 | 0.091 | lig1 | -0.052 |
| rtn1b | 0.091 | rrm2 | -0.052 |