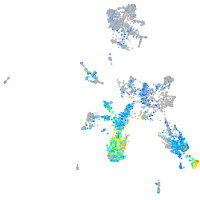
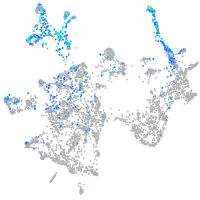
calcyphosine-like a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.471 | ahcy | -0.143 |
| insm1b | 0.436 | gamt | -0.139 |
| c2cd4a | 0.411 | gapdh | -0.136 |
| neurod1 | 0.400 | hdlbpa | -0.133 |
| egr4 | 0.397 | fabp3 | -0.133 |
| zgc:101731 | 0.379 | gatm | -0.127 |
| fev | 0.372 | bhmt | -0.123 |
| pacrg | 0.365 | fbp1b | -0.118 |
| mir7a-1 | 0.360 | mat1a | -0.116 |
| pcsk1nl | 0.358 | aqp12 | -0.113 |
| arxa | 0.357 | nupr1b | -0.111 |
| hepacam2 | 0.356 | aldh6a1 | -0.110 |
| mir375-2 | 0.353 | rpl6 | -0.110 |
| dll4 | 0.348 | apoa4b.1 | -0.108 |
| hsbp1a | 0.346 | apoa1b | -0.107 |
| pih1d3 | 0.344 | cx32.3 | -0.107 |
| tmsb | 0.341 | afp4 | -0.106 |
| capslb | 0.341 | apoc1 | -0.106 |
| si:dkey-83m22.7 | 0.340 | rpl4 | -0.105 |
| ccka | 0.339 | gstt1a | -0.105 |
| si:ch73-160i9.2 | 0.335 | si:dkey-16p21.8 | -0.103 |
| zgc:195356 | 0.333 | apoc2 | -0.103 |
| rasd4 | 0.332 | agxtb | -0.103 |
| ccdc173 | 0.331 | aldh7a1 | -0.102 |
| TCIM (1 of many) | 0.329 | gnmt | -0.102 |
| meig1 | 0.322 | hspd1 | -0.100 |
| id4 | 0.317 | eif4ebp1 | -0.100 |
| vat1 | 0.316 | shmt1 | -0.100 |
| si:ch211-147d7.5 | 0.313 | hspe1 | -0.099 |
| chga | 0.313 | tfa | -0.097 |
| tekt1 | 0.311 | mgst1.2 | -0.097 |
| nkx2.2a | 0.309 | sec61a1 | -0.097 |
| enkur | 0.309 | dap | -0.096 |
| spata18 | 0.308 | scp2a | -0.095 |
| zgc:165481 | 0.305 | serpina1 | -0.095 |