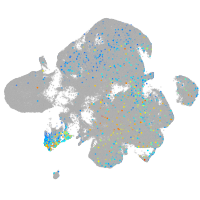
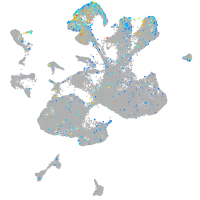
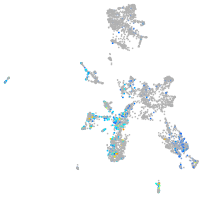
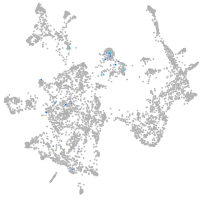
"calpain 1, (mu/I) large subunit b"
ZFIN 
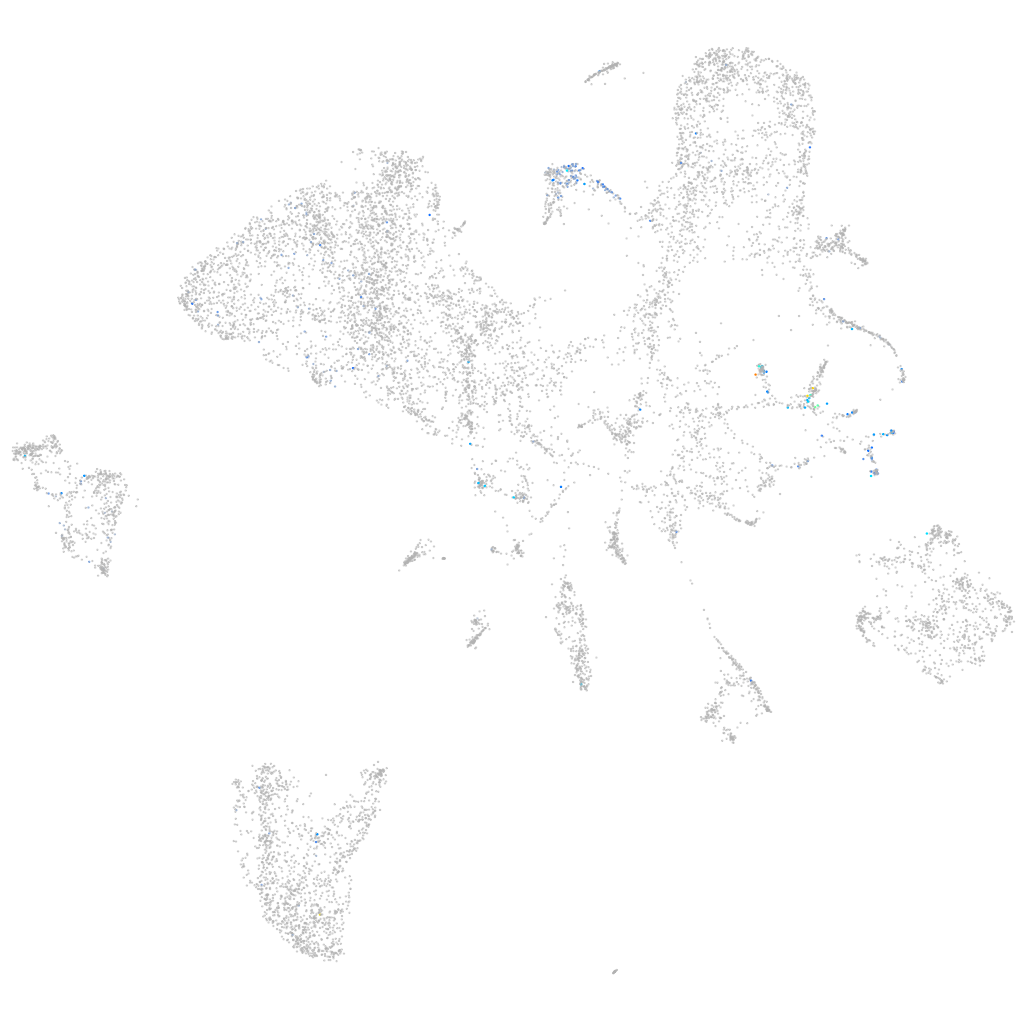
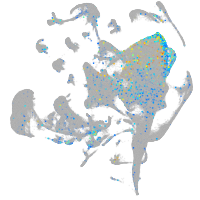
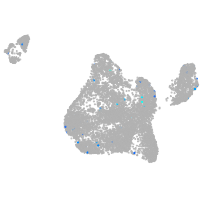
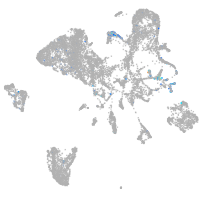
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110438234 | 0.253 | ahcy | -0.082 |
| si:ch211-163m17.4 | 0.201 | gapdh | -0.074 |
| scg2b | 0.190 | gamt | -0.066 |
| nsfa | 0.185 | gatm | -0.064 |
| XLOC-013106 | 0.183 | hspe1 | -0.064 |
| arhgap22 | 0.180 | fbp1b | -0.062 |
| scg3 | 0.177 | nupr1b | -0.061 |
| neurod1 | 0.176 | si:dkey-16p21.8 | -0.061 |
| id4 | 0.175 | gstt1a | -0.060 |
| calca | 0.173 | gstr | -0.059 |
| insm1b | 0.170 | apoc2 | -0.059 |
| rem2 | 0.169 | bhmt | -0.059 |
| AL935044.4 | 0.166 | npm1a | -0.059 |
| vat1 | 0.165 | apoa4b.1 | -0.058 |
| LOC110439873 | 0.164 | glud1b | -0.058 |
| fam167ab | 0.163 | rps18 | -0.056 |
| pcsk1nl | 0.162 | aldh6a1 | -0.056 |
| nalcn | 0.161 | aldh7a1 | -0.056 |
| cpeb1b | 0.161 | mat1a | -0.056 |
| otogl | 0.160 | apoa1b | -0.055 |
| LOC100331291 | 0.159 | scp2a | -0.055 |
| BX950188.1 | 0.159 | eif4ebp1 | -0.054 |
| CR556712.1 | 0.159 | aqp12 | -0.054 |
| tspan7b | 0.158 | hspd1 | -0.054 |
| anxa5a | 0.158 | pnp4b | -0.053 |
| aldocb | 0.158 | ssr1 | -0.053 |
| calm1b | 0.158 | sult2st2 | -0.053 |
| tmsb | 0.157 | rgn | -0.052 |
| gapdhs | 0.156 | dap | -0.052 |
| capn2b | 0.155 | suclg1 | -0.052 |
| kcnc1b | 0.155 | eif4bb | -0.051 |
| nkx2.2a | 0.155 | fabp3 | -0.051 |
| mir7a-1 | 0.154 | suclg2 | -0.051 |
| cdh26.2 | 0.154 | agxta | -0.050 |
| foxq1b | 0.154 | gcshb | -0.050 |