"calmodulin regulated spectrin-associated protein family, member 2a"
ZFIN 
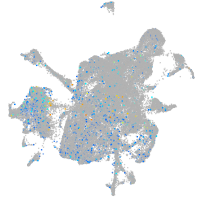
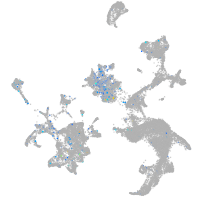
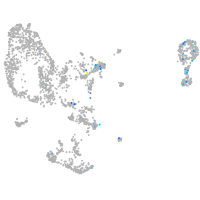
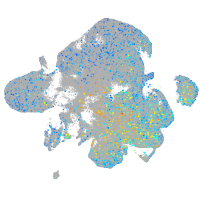
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.195 | id1 | -0.134 |
| elavl4 | 0.184 | mdka | -0.120 |
| stx1b | 0.183 | XLOC-003690 | -0.116 |
| myt1b | 0.182 | sparc | -0.103 |
| ywhah | 0.175 | vamp3 | -0.103 |
| syt2a | 0.175 | atp1a1b | -0.100 |
| cplx2 | 0.172 | fabp7a | -0.099 |
| snap25a | 0.171 | cldn5a | -0.099 |
| rbfox1 | 0.171 | sdc4 | -0.096 |
| sncb | 0.169 | her15.1 | -0.094 |
| stxbp1a | 0.168 | cd63 | -0.093 |
| gng3 | 0.167 | sox3 | -0.092 |
| ywhag2 | 0.162 | COX7A2 (1 of many) | -0.092 |
| rtn1b | 0.158 | cd82a | -0.090 |
| gng2 | 0.155 | hmgb2a | -0.089 |
| tmem59l | 0.154 | si:ch211-286b5.5 | -0.089 |
| stmn2a | 0.154 | fosab | -0.089 |
| kcnc3a | 0.153 | qki2 | -0.088 |
| atp1b2a | 0.153 | msi1 | -0.088 |
| stmn1b | 0.153 | msna | -0.087 |
| vamp2 | 0.153 | selenoh | -0.087 |
| onecut1 | 0.152 | pgrmc1 | -0.084 |
| atp6v0cb | 0.152 | pcna | -0.084 |
| rtn1a | 0.151 | her9 | -0.084 |
| gabrb4 | 0.151 | sox2 | -0.082 |
| atp1a3a | 0.151 | sox19a | -0.081 |
| cplx2l | 0.151 | zgc:56493 | -0.080 |
| tuba2 | 0.149 | ccng1 | -0.080 |
| aplp1 | 0.149 | dut | -0.080 |
| gap43 | 0.148 | btg2 | -0.079 |
| golga7ba | 0.147 | endouc | -0.079 |
| id4 | 0.147 | si:ch73-265d7.2 | -0.079 |
| tubb5 | 0.147 | her12 | -0.078 |
| stmn2b | 0.145 | psat1 | -0.077 |
| nova1 | 0.145 | notch3 | -0.076 |