Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 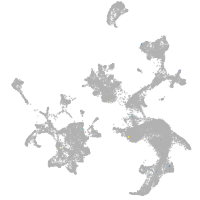 |
pronephros  |
neural |
spinal cord / glia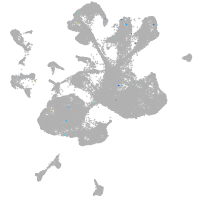 |
eye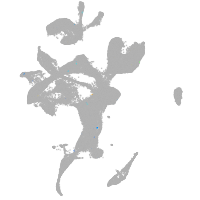 |
taste / olfactory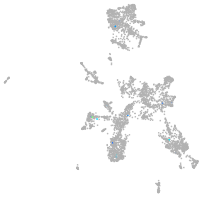 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-043859 | 0.359 | rps26 | -0.036 |
| zgc:194551 | 0.308 | rpl22 | -0.031 |
| ccdc113 | 0.262 | rpl37 | -0.031 |
| arhgef28 | 0.182 | rpl14 | -0.031 |
| si:ch211-195j11.30 | 0.180 | serbp1a | -0.027 |
| vegfd | 0.178 | actb2 | -0.026 |
| h6pd | 0.172 | mt-cyb | -0.026 |
| aplnr2 | 0.164 | mt-nd1 | -0.026 |
| creld1a | 0.161 | rpl26 | -0.025 |
| prmt9 | 0.160 | pabpc1a | -0.025 |
| znf1092 | 0.148 | cotl1 | -0.022 |
| CR848736.1 | 0.139 | rpl31 | -0.022 |
| tamm41 | 0.136 | hbbe1.1 | -0.019 |
| si:ch73-72b7.1 | 0.135 | uqcrq | -0.018 |
| ryr1a | 0.134 | uba52 | -0.018 |
| trim65 | 0.130 | mt-nd3 | -0.018 |
| ptpn23a | 0.129 | rsl24d1 | -0.018 |
| rnf24 | 0.128 | ap2m1a | -0.018 |
| snai1b | 0.123 | ndufb8 | -0.018 |
| tbce | 0.118 | gnb1a | -0.018 |
| pcolce2a | 0.118 | tfap2e | -0.018 |
| arfgap3 | 0.115 | canx | -0.018 |
| gucy2c | 0.114 | rab38 | -0.018 |
| KCNIP4 | 0.113 | hbae1.1 | -0.017 |
| bean1 | 0.113 | rplp0 | -0.017 |
| tmem45b | 0.113 | mt-co2 | -0.017 |
| cyp27a7 | 0.111 | ndufb3 | -0.017 |
| lhfpl6 | 0.111 | rplp2l | -0.017 |
| n4bp2 | 0.110 | rpl27a | -0.017 |
| psmd5 | 0.110 | zgc:165573 | -0.017 |
| dhrsx | 0.107 | rps18 | -0.017 |
| pvalb3 | 0.107 | hmgn7 | -0.017 |
| prdm8b | 0.107 | pttg1ipb | -0.017 |
| nlrx1 | 0.099 | tma7 | -0.017 |
| rbfox1l | 0.099 | blcap | -0.017 |