Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia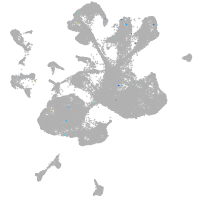 |
eye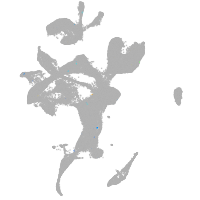 |
taste / olfactory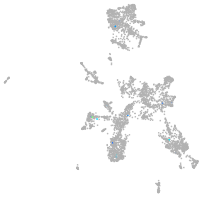 |
otic / lateral line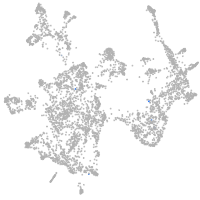 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ggact.3 | 0.148 | rpl24 | -0.023 |
| LOC100534673 | 0.101 | serbp1a | -0.022 |
| glipr2 | 0.093 | rpl36a | -0.022 |
| LOC101885801 | 0.091 | rps20 | -0.020 |
| XLOC-028682 | 0.090 | rpl39 | -0.020 |
| alpk3a | 0.085 | rpl15 | -0.019 |
| dapk2a | 0.084 | rpl31 | -0.019 |
| BX649490.2 | 0.078 | anp32a | -0.019 |
| g6pcb | 0.078 | aldoaa | -0.019 |
| eps8l3b | 0.075 | rpl10 | -0.018 |
| anpepb | 0.071 | cnbpb | -0.018 |
| CR376800.1 | 0.069 | drap1 | -0.018 |
| tspan10 | 0.068 | zgc:114188 | -0.018 |
| CU062645.1 | 0.067 | atp5mc3a | -0.018 |
| BX005054.1 | 0.064 | btf3 | -0.018 |
| XLOC-012657 | 0.063 | sap18 | -0.018 |
| mir430b-10 | 0.063 | rsl24d1 | -0.017 |
| ptgis | 0.061 | eif3k | -0.017 |
| sgsm1b | 0.060 | setb | -0.017 |
| si:dkey-28d5.14 | 0.059 | rps12 | -0.017 |
| CU459094.2 | 0.058 | rps21 | -0.017 |
| traf3ip2l | 0.057 | tma7 | -0.017 |
| chrnb2l | 0.057 | rack1 | -0.017 |
| tnni1d | 0.056 | hmga1a | -0.017 |
| klc3 | 0.055 | rps26 | -0.017 |
| CR387932.1 | 0.055 | rpl28 | -0.017 |
| si:ch211-284e13.14 | 0.054 | rpl12 | -0.016 |
| LOC103910489 | 0.052 | rps10 | -0.016 |
| ikzf2 | 0.052 | rpl36 | -0.016 |
| gngt1 | 0.051 | rpl22 | -0.016 |
| pdcb | 0.051 | rpl8 | -0.016 |
| RF00611 | 0.050 | gtpbp4 | -0.016 |
| XLOC-002411 | 0.049 | hmgn2 | -0.016 |
| il12ba | 0.048 | eif3i | -0.016 |
| XLOC-024828 | 0.047 | btg2 | -0.016 |




