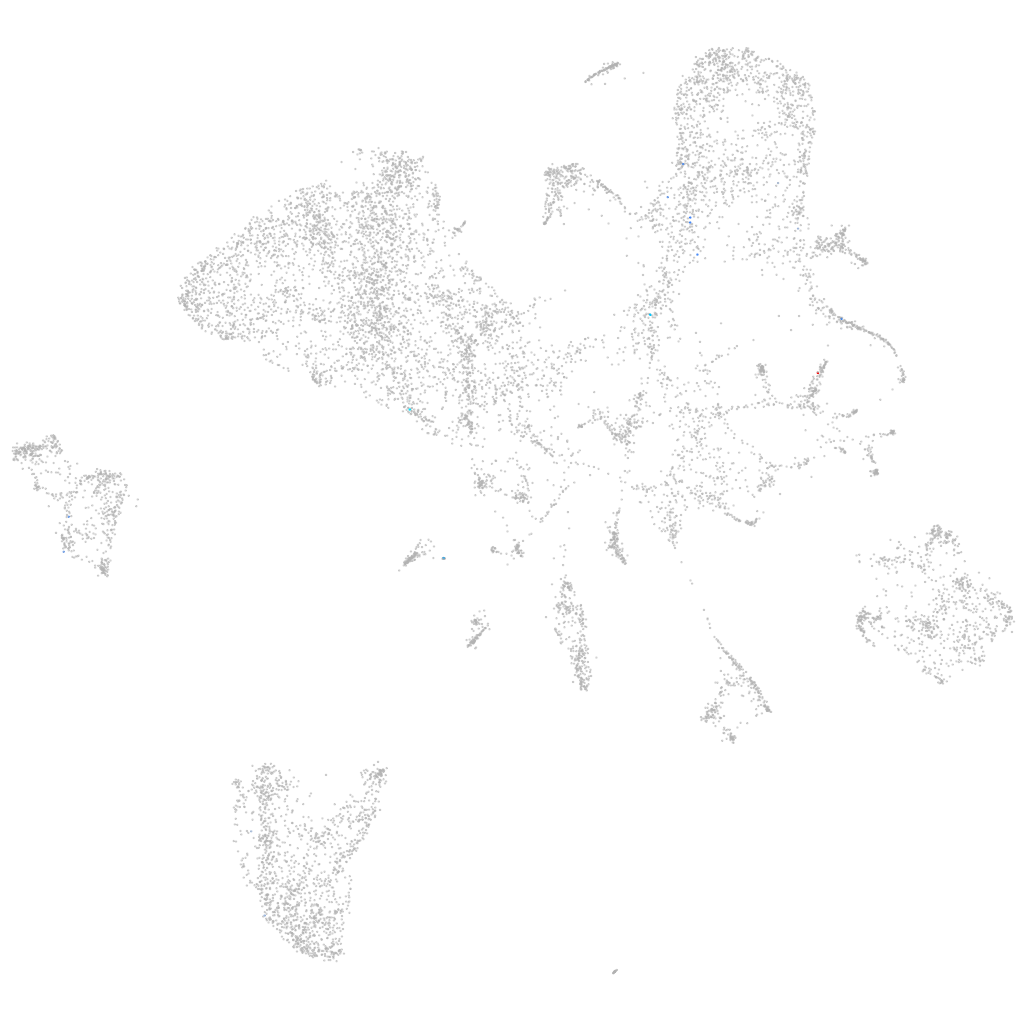
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| avpr1ab | 0.628 | ybx1 | -0.025 |
| wdr78 | 0.575 | rps12 | -0.021 |
| si:dkey-7i4.5 | 0.543 | rpl5b | -0.020 |
| rxfp3.2a | 0.361 | rps18 | -0.018 |
| lrfn2b | 0.337 | bhmt | -0.017 |
| pfkmb | 0.300 | pnrc2 | -0.017 |
| si:dkey-222h21.2 | 0.283 | sec61a1 | -0.015 |
| ccdc181 | 0.257 | rpl22l1 | -0.015 |
| si:dkey-181m9.8 | 0.252 | si:dkey-56m19.5 | -0.015 |
| si:dkey-216e24.9 | 0.228 | gatm | -0.015 |
| mapta | 0.224 | hgd | -0.015 |
| kif21a | 0.206 | igfbp2a | -0.015 |
| CABZ01034698.2 | 0.201 | gpd1b | -0.015 |
| plch1 | 0.189 | aqp12 | -0.014 |
| cers6 | 0.185 | hal | -0.014 |
| CU571064.1 | 0.181 | si:dkey-16p21.8 | -0.014 |
| st3gal2 | 0.175 | proca | -0.014 |
| egr2b | 0.172 | hmgn7 | -0.014 |
| cacna1ha | 0.168 | sdc2 | -0.014 |
| itga6a | 0.168 | f7 | -0.014 |
| gpd2 | 0.166 | shmt1 | -0.014 |
| hdac11 | 0.156 | agla | -0.013 |
| LOC108191602 | 0.153 | tfa | -0.013 |
| mcoln1a | 0.152 | cpn1 | -0.013 |
| hmbsb | 0.150 | si:ch1073-325m22.2 | -0.013 |
| malt3 | 0.150 | ttc36 | -0.013 |
| pgbd5 | 0.147 | gamt | -0.013 |
| micall2b | 0.145 | allc | -0.013 |
| bin1a | 0.144 | dgat2 | -0.013 |
| dock4b | 0.143 | pcbd1 | -0.013 |
| armc8 | 0.140 | apoc1 | -0.013 |
| atg4da | 0.137 | apoa1b | -0.013 |
| si:dkey-44g17.6 | 0.137 | qdpra | -0.013 |
| cetn2 | 0.136 | fabp3 | -0.013 |
| lrrc4ba | 0.135 | hvcn1 | -0.013 |