Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye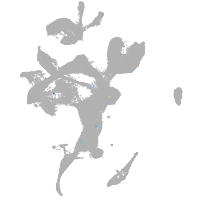 |
taste / olfactory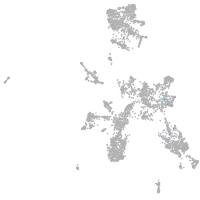 |
otic / lateral line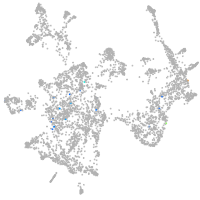 |
epidermis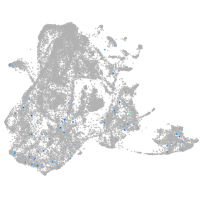 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU019662.1 | 0.034 | actc1b | -0.013 |
| si:dkey-185m8.2 | 0.034 | elavl3 | -0.013 |
| cks1b | 0.033 | rtn1a | -0.013 |
| si:ch211-214p16.1 | 0.033 | gpm6aa | -0.012 |
| si:ch73-208g10.1 | 0.032 | gpm6ab | -0.012 |
| tuba8l | 0.032 | pvalb2 | -0.012 |
| cdk1 | 0.031 | stmn1b | -0.012 |
| laptm5 | 0.031 | ckbb | -0.011 |
| lcp1 | 0.031 | pvalb1 | -0.011 |
| si:dkeyp-68b7.12 | 0.031 | tmsb | -0.011 |
| wasa | 0.031 | hbbe1.3 | -0.010 |
| aurkb | 0.030 | myt1a | -0.010 |
| rasal3 | 0.030 | myt1b | -0.010 |
| lpar5a | 0.030 | scrt2 | -0.010 |
| lpxn | 0.030 | dpysl3 | -0.009 |
| BX927258.1 | 0.029 | gng3 | -0.009 |
| coro1a | 0.029 | mylpfa | -0.009 |
| dut | 0.029 | nova2 | -0.009 |
| grap2a | 0.029 | atp6v0cb | -0.008 |
| mad2l1 | 0.029 | celf3a | -0.008 |
| FERMT3 (1 of many) | 0.029 | hbae3 | -0.008 |
| CU499330.1 | 0.028 | LOC798783 | -0.008 |
| gpr183a | 0.028 | rnasekb | -0.008 |
| grap2b | 0.028 | rtn1b | -0.008 |
| itgae.2 | 0.028 | si:dkey-56m19.5 | -0.008 |
| nutf2l | 0.028 | si:dkeyp-75h12.5 | -0.008 |
| unc13d | 0.028 | sncb | -0.008 |
| XLOC-039121 | 0.028 | tpma | -0.008 |
| zgc:56493 | 0.028 | tuba1c | -0.008 |
| cdc20 | 0.027 | ube2d4 | -0.008 |
| cdca5 | 0.027 | ebf3a | -0.008 |
| cxcr3.2 | 0.027 | acta1b | -0.007 |
| glipr1a | 0.027 | cadm3 | -0.007 |
| kpna2 | 0.027 | csdc2a | -0.007 |
| mki67 | 0.027 | dbn1 | -0.007 |




