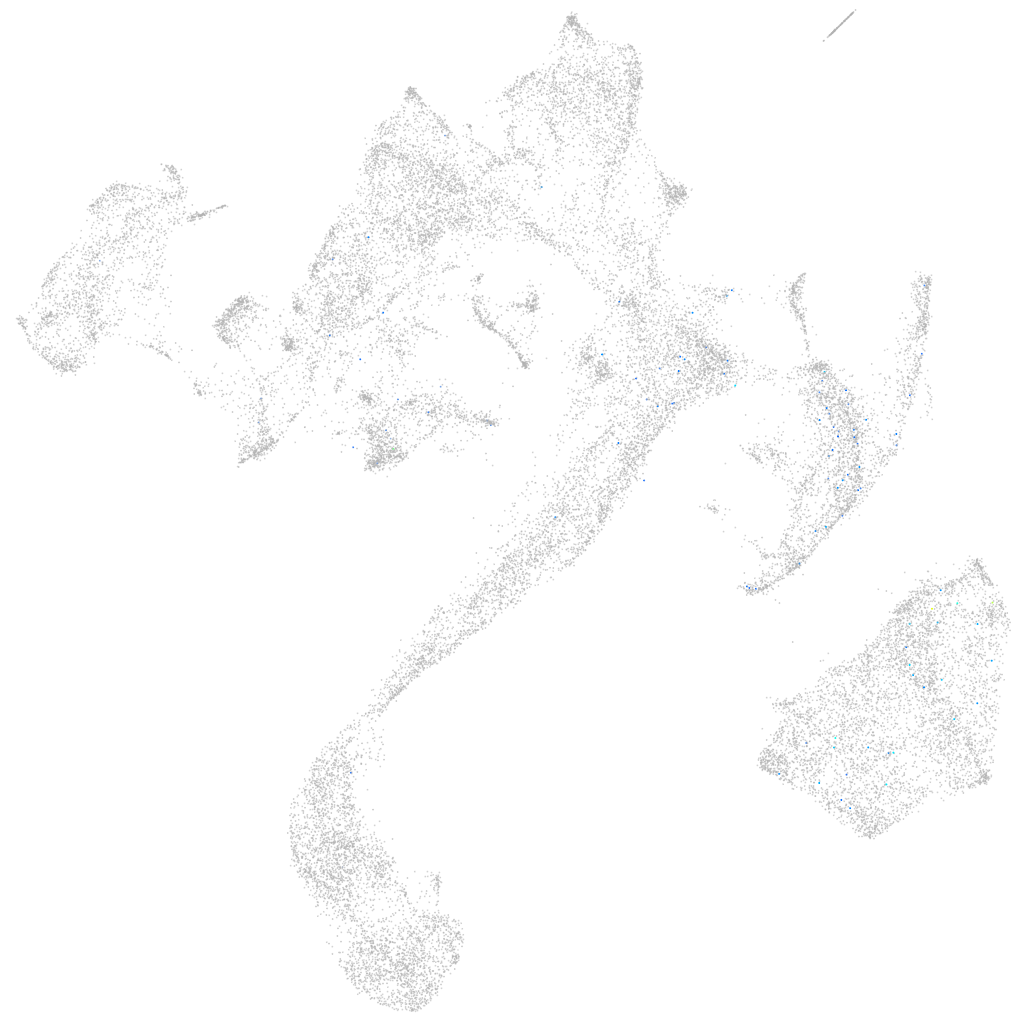
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX957362.5 | 0.142 | fabp3 | -0.037 |
| CU467880.1 | 0.140 | actc1b | -0.035 |
| XLOC-037784 | 0.135 | tpi1b | -0.030 |
| LOC100006699 | 0.124 | ak1 | -0.030 |
| BX530024.1 | 0.119 | gamt | -0.030 |
| EIF3J | 0.118 | ckmb | -0.029 |
| BX901962.4 | 0.113 | bhmt | -0.029 |
| LO017820.1 | 0.105 | ckma | -0.029 |
| tmem119a | 0.103 | atp2a1 | -0.029 |
| BX510949.1 | 0.102 | aldoab | -0.029 |
| BX000981.5 | 0.099 | pabpc4 | -0.029 |
| XLOC-016411 | 0.097 | tpma | -0.029 |
| LOC103910876 | 0.096 | eno1a | -0.028 |
| AL935198.2 | 0.091 | acta1b | -0.028 |
| pimr196 | 0.086 | sparc | -0.028 |
| shpk | 0.084 | mylpfa | -0.028 |
| kcnj9 | 0.081 | tnnc2 | -0.028 |
| rinl | 0.076 | eno3 | -0.027 |
| BX323459.2 | 0.076 | atp5meb | -0.027 |
| nts | 0.076 | nme2b.2 | -0.027 |
| LOC110438267 | 0.074 | rplp2 | -0.027 |
| AL627126.1 | 0.072 | mdh2 | -0.027 |
| CR854988.1 | 0.070 | fxyd6l | -0.027 |
| LOC103909745 | 0.069 | neb | -0.027 |
| FP089511.1 | 0.068 | ldb3b | -0.026 |
| si:dkey-30j10.5 | 0.068 | ldb3a | -0.026 |
| si:dkey-187j14.6 | 0.067 | tmem38a | -0.026 |
| LOC103910259 | 0.067 | eef1da | -0.026 |
| si:dkey-220o21.3 | 0.067 | CABZ01078594.1 | -0.026 |
| XLOC-038929 | 0.064 | vdac3 | -0.026 |
| RF00324 | 0.063 | ttn.2 | -0.026 |
| shisa6 | 0.062 | srl | -0.026 |
| BX293991.1 | 0.061 | gapdh | -0.026 |
| XLOC-011600 | 0.060 | col1a1a | -0.026 |
| XLOC-036658 | 0.060 | mylz3 | -0.026 |