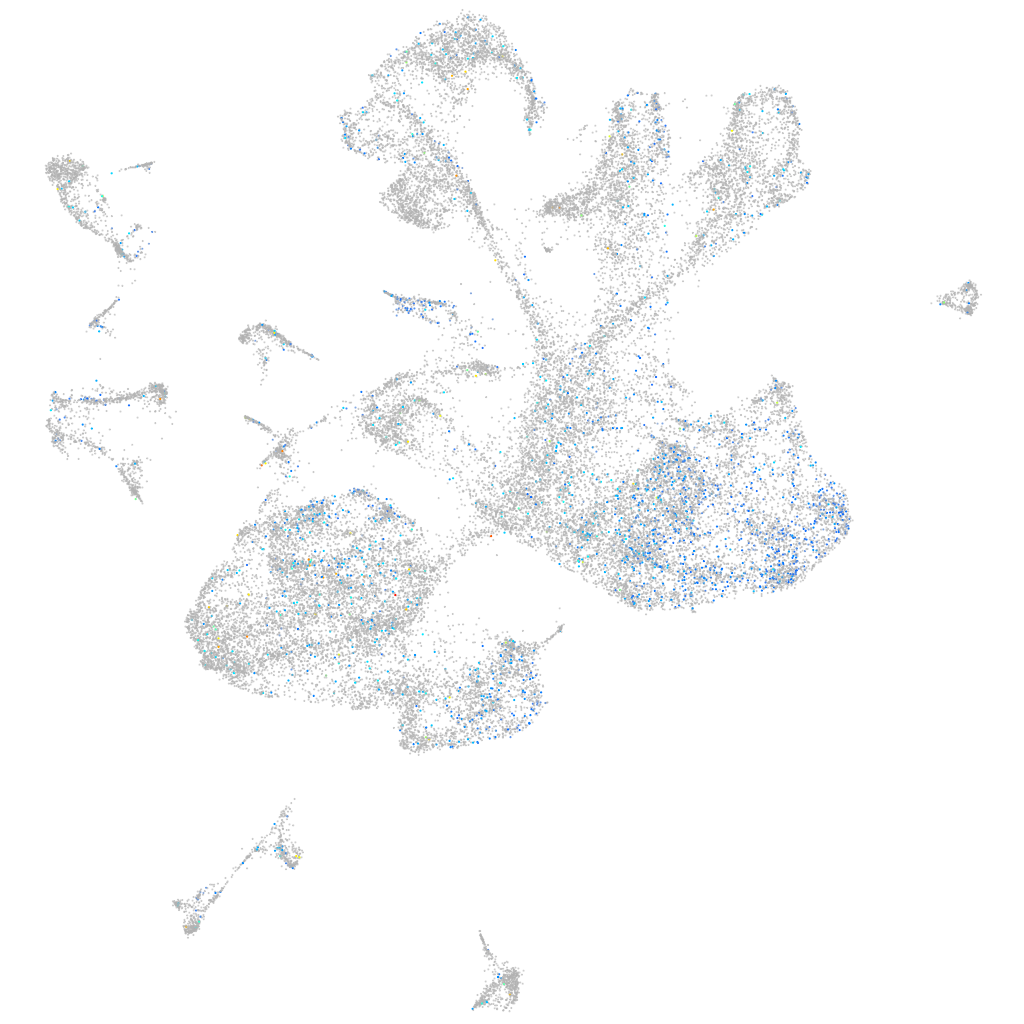
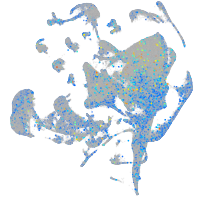
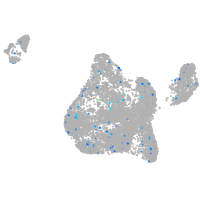
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.107 | CR383676.1 | -0.063 |
| dut | 0.106 | myt1b | -0.055 |
| rrm1 | 0.104 | rtn1a | -0.054 |
| tuba8l4 | 0.103 | ppdpfb | -0.053 |
| rpa3 | 0.097 | gpm6ab | -0.051 |
| nutf2l | 0.095 | ckbb | -0.051 |
| fen1 | 0.095 | elavl3 | -0.051 |
| ranbp1 | 0.093 | gpm6aa | -0.050 |
| stmn1a | 0.093 | si:ch211-260e23.9 | -0.049 |
| nasp | 0.093 | pik3r3b | -0.047 |
| banf1 | 0.092 | si:dkeyp-75h12.5 | -0.044 |
| anp32b | 0.091 | pvalb1 | -0.042 |
| rpa2 | 0.091 | nanos1 | -0.041 |
| rnaseh2a | 0.091 | stmn1b | -0.041 |
| hmgb2a | 0.090 | serinc1 | -0.041 |
| cbx3a | 0.090 | tp53inp1 | -0.040 |
| sumo3b | 0.089 | abcc5 | -0.040 |
| npm1a | 0.089 | rnasekb | -0.040 |
| nop58 | 0.089 | sox4a | -0.039 |
| cnbpa | 0.088 | LOC100537384 | -0.039 |
| cks1b | 0.088 | gng3 | -0.038 |
| chaf1a | 0.088 | COX3 | -0.038 |
| snrpd1 | 0.088 | sesn1 | -0.037 |
| selenoh | 0.087 | pvalb2 | -0.037 |
| pa2g4a | 0.087 | vamp2 | -0.037 |
| mcm7 | 0.087 | znf704 | -0.037 |
| shmt1 | 0.087 | ube2d4 | -0.037 |
| zgc:56493 | 0.086 | marcksl1b | -0.037 |
| dek | 0.086 | mkrn1 | -0.036 |
| snrpe | 0.085 | elavl4 | -0.036 |
| snrpf | 0.084 | sncb | -0.036 |
| dkc1 | 0.083 | h1f0 | -0.036 |
| ssrp1a | 0.083 | gabarapa | -0.036 |
| snu13b | 0.083 | stx1b | -0.036 |
| hmgb2b | 0.083 | calm1a | -0.036 |