Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye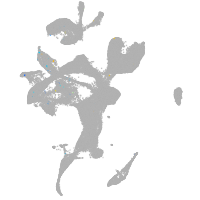 |
taste / olfactory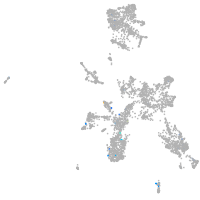 |
otic / lateral line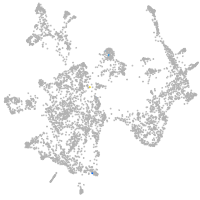 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells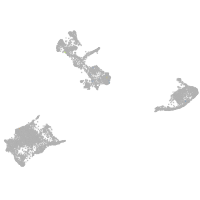 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103911745 | 0.110 | hmgb2a | -0.042 |
| LOC103908977 | 0.104 | mdka | -0.028 |
| rbpms2a | 0.095 | dut | -0.028 |
| inab | 0.080 | rrm1 | -0.027 |
| CU856621.2 | 0.076 | ahcy | -0.027 |
| pou4f1 | 0.075 | pcna | -0.026 |
| BX005073.1 | 0.073 | msi1 | -0.026 |
| pou4f3 | 0.073 | si:ch73-281n10.2 | -0.024 |
| scn8aa | 0.072 | eef1da | -0.024 |
| rbpms2b | 0.071 | cks1b | -0.024 |
| pou4f2 | 0.069 | COX7A2 (1 of many) | -0.024 |
| nrn1a | 0.068 | lbr | -0.024 |
| FAM163B (1 of many) | 0.068 | stmn1a | -0.024 |
| isl2b | 0.068 | anp32b | -0.023 |
| BX247868.1 | 0.066 | mki67 | -0.023 |
| stmn2a | 0.066 | hmga1a | -0.023 |
| lct | 0.065 | psat1 | -0.023 |
| kcnq2a | 0.064 | chaf1a | -0.023 |
| eno2 | 0.063 | nutf2l | -0.023 |
| stmn4 | 0.063 | ccng1 | -0.023 |
| LOC110438169 | 0.062 | mcm7 | -0.023 |
| maptb | 0.062 | selenoh | -0.023 |
| rtn1b | 0.062 | tuba8l | -0.023 |
| LOC108179130 | 0.062 | msna | -0.023 |
| BX323830.1 | 0.061 | dek | -0.022 |
| syt2a | 0.061 | pa2g4b | -0.022 |
| gria4a | 0.061 | mibp | -0.022 |
| elavl3 | 0.060 | ccna2 | -0.022 |
| ywhag1 | 0.060 | nasp | -0.022 |
| zgc:158291 | 0.060 | lig1 | -0.022 |
| LO018598.1 | 0.060 | ncapg | -0.022 |
| vsnl1b | 0.059 | si:ch211-156b7.4 | -0.022 |
| cplx2 | 0.059 | atp5mc3b | -0.021 |
| gng3 | 0.059 | her15.1 | -0.021 |
| tubb5 | 0.058 | rpa3 | -0.021 |




