Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 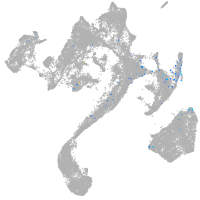 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 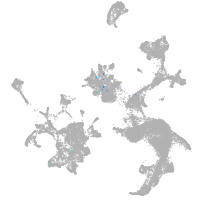 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells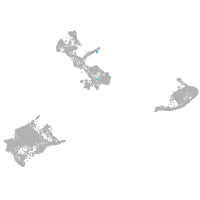 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cspg5a | 0.042 | aldob | -0.024 |
| atp6v0cb | 0.040 | epcam | -0.023 |
| sncb | 0.040 | cd9b | -0.021 |
| ywhag2 | 0.038 | cfl1l | -0.021 |
| elavl4 | 0.037 | hdlbpa | -0.021 |
| rtn1b | 0.037 | eef1da | -0.020 |
| stmn2a | 0.037 | f11r.1 | -0.020 |
| tbx6 | 0.037 | gstp1 | -0.020 |
| gap43 | 0.036 | krt4 | -0.020 |
| gng3 | 0.036 | s100a10b | -0.020 |
| vamp2 | 0.036 | wu:fb18f06 | -0.020 |
| zgc:65894 | 0.036 | zgc:175088 | -0.020 |
| gpm6aa | 0.034 | anxa1a | -0.019 |
| sv2a | 0.034 | capns1a | -0.019 |
| sypb | 0.034 | cast | -0.019 |
| basp1 | 0.033 | cyt1 | -0.019 |
| myt1la | 0.033 | icn | -0.019 |
| rtn1a | 0.033 | spint2 | -0.019 |
| tuba1c | 0.033 | cdh1 | -0.018 |
| syt1a | 0.033 | cldnb | -0.018 |
| atpv0e2 | 0.032 | cst14b.1 | -0.018 |
| camk2d2 | 0.032 | cyt1l | -0.018 |
| celf5a | 0.032 | gsta.1 | -0.018 |
| map1aa | 0.032 | icn2 | -0.018 |
| nrxn1a | 0.032 | krt5 | -0.018 |
| nsg2 | 0.032 | krt8 | -0.018 |
| rnasekb | 0.032 | lye | -0.018 |
| si:dkeyp-75h12.5 | 0.032 | oclna | -0.018 |
| snap25a | 0.032 | pfn1 | -0.018 |
| sprn | 0.032 | pycard | -0.018 |
| stx1b | 0.032 | si:ch211-195b11.3 | -0.018 |
| syngr3a | 0.032 | sult6b1 | -0.018 |
| gabrb4 | 0.032 | zgc:153284 | -0.018 |
| stxbp1a | 0.032 | zgc:162730 | -0.018 |
| aplp1 | 0.031 | anxa1c | -0.017 |




