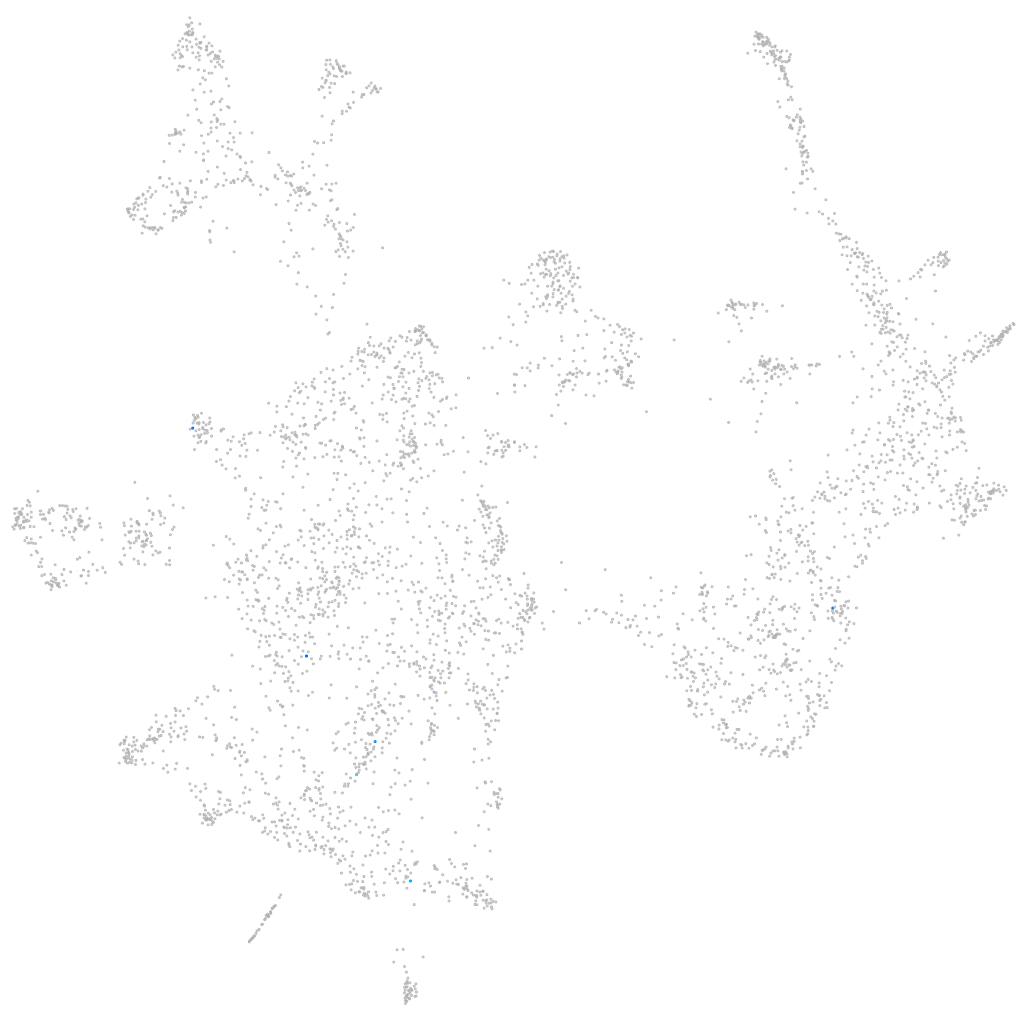
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mcf2b | 0.533 | si:dkey-222f2.1 | -0.039 |
| rcvrnb | 0.433 | atp5mf | -0.035 |
| XLOC-018188 | 0.413 | irf2bp2b | -0.031 |
| trnW | 0.409 | mrpl36 | -0.030 |
| XLOC-012207 | 0.298 | cycsb | -0.029 |
| si:dkey-193b15.5 | 0.294 | rnf185 | -0.029 |
| LOC110437824 | 0.283 | chrac1 | -0.029 |
| LOC101885894 | 0.276 | tuba8l | -0.028 |
| XLOC-002665 | 0.271 | pvalb2 | -0.028 |
| pimr184 | 0.245 | anp32e | -0.028 |
| RAMP3 | 0.232 | minos1 | -0.027 |
| XLOC-027769 | 0.207 | ywhaqa | -0.027 |
| baalca | 0.203 | eya1 | -0.027 |
| OSCP1 (1 of many) | 0.193 | rarga | -0.027 |
| c3a.6 | 0.188 | hspa4b | -0.027 |
| zgc:172051 | 0.187 | atp5mc1 | -0.027 |
| cnr1 | 0.175 | calm1a | -0.026 |
| BX664614.1 | 0.171 | mrps14 | -0.026 |
| calcoco2 | 0.167 | psmd6 | -0.026 |
| timp4.1 | 0.165 | ndufb7 | -0.026 |
| klhdc8a | 0.163 | mrps34 | -0.025 |
| sfrp1b | 0.149 | map1lc3b | -0.025 |
| mrtfaa | 0.148 | mt-nd4 | -0.025 |
| mylk3 | 0.146 | gapdhs | -0.025 |
| cmtm7 | 0.145 | cdkn1ca | -0.025 |
| TMCC1 | 0.142 | cfl1 | -0.025 |
| LOC101885990 | 0.142 | dpm2 | -0.024 |
| pde11a | 0.141 | mrps18c | -0.024 |
| zgc:171727 | 0.134 | ndufb10 | -0.024 |
| zgc:110348 | 0.131 | ufm1 | -0.024 |
| zgc:136908 | 0.131 | kif5ba | -0.024 |
| tmtops3a | 0.128 | tomm40 | -0.024 |
| CABZ01067348.1 | 0.127 | eya4 | -0.024 |
| si:ch73-44m9.2 | 0.127 | pde4ba | -0.024 |
| XLOC-036929 | 0.126 | trappc3 | -0.024 |