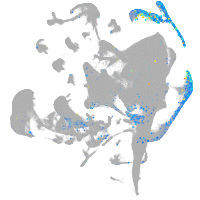
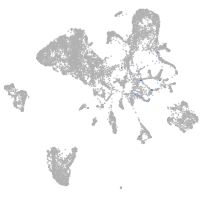
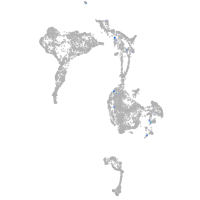
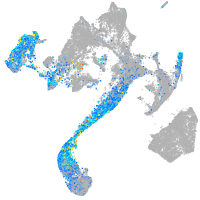
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mamdc2a | 0.406 | cox8a | -0.051 |
| CT574585.1 | 0.294 | zgc:158463 | -0.039 |
| nphs2 | 0.292 | mdh1aa | -0.038 |
| si:dkey-27l15.1 | 0.270 | sod1 | -0.038 |
| CT027772.2 | 0.258 | nipsnap2 | -0.037 |
| b3gnt9 | 0.249 | mt-nd5 | -0.036 |
| si:dkey-238k10.3 | 0.245 | ndufb2 | -0.036 |
| hephl1b | 0.244 | atp2b1a | -0.036 |
| CABZ01043953.1.1 | 0.208 | chga | -0.035 |
| zgc:154086 | 0.205 | COX3 | -0.033 |
| xkr5a | 0.204 | chmp2a | -0.031 |
| NC-002333.20 | 0.190 | htatip2 | -0.031 |
| CT027756.1 | 0.188 | fstl5 | -0.031 |
| BX936386.1 | 0.168 | sdcbp2 | -0.030 |
| LOC100002202 | 0.168 | wu:fb18f06 | -0.030 |
| slbp2 | 0.165 | clta | -0.030 |
| pvalb3 | 0.160 | ankrd12 | -0.030 |
| CR927456.1 | 0.155 | mt-co2 | -0.029 |
| AL953907.2 | 0.153 | gstp1 | -0.029 |
| nos1 | 0.149 | eno1a | -0.029 |
| BX548061.1 | 0.148 | gapdhs | -0.029 |
| dlc | 0.148 | nqo1 | -0.028 |
| wu:fj20b03 | 0.145 | ndufs4 | -0.028 |
| srl | 0.144 | fxyd1 | -0.028 |
| XLOC-005272 | 0.142 | mtdha | -0.028 |
| XLOC-031633 | 0.141 | map1aa | -0.028 |
| he1.1 | 0.140 | oclna | -0.028 |
| BX470188.1 | 0.139 | kif5bb | -0.028 |
| LOC110437784 | 0.137 | serinc2 | -0.028 |
| si:dkey-159n16.2 | 0.137 | sdhc | -0.028 |
| LOC103910002 | 0.133 | mt-atp6 | -0.028 |
| lrrc3cb | 0.133 | hnrpkl | -0.028 |
| ccdc106b | 0.133 | tusc3 | -0.028 |
| XLOC-041707 | 0.131 | tmed7 | -0.028 |
| AL645691.1 | 0.130 | atf4a | -0.028 |