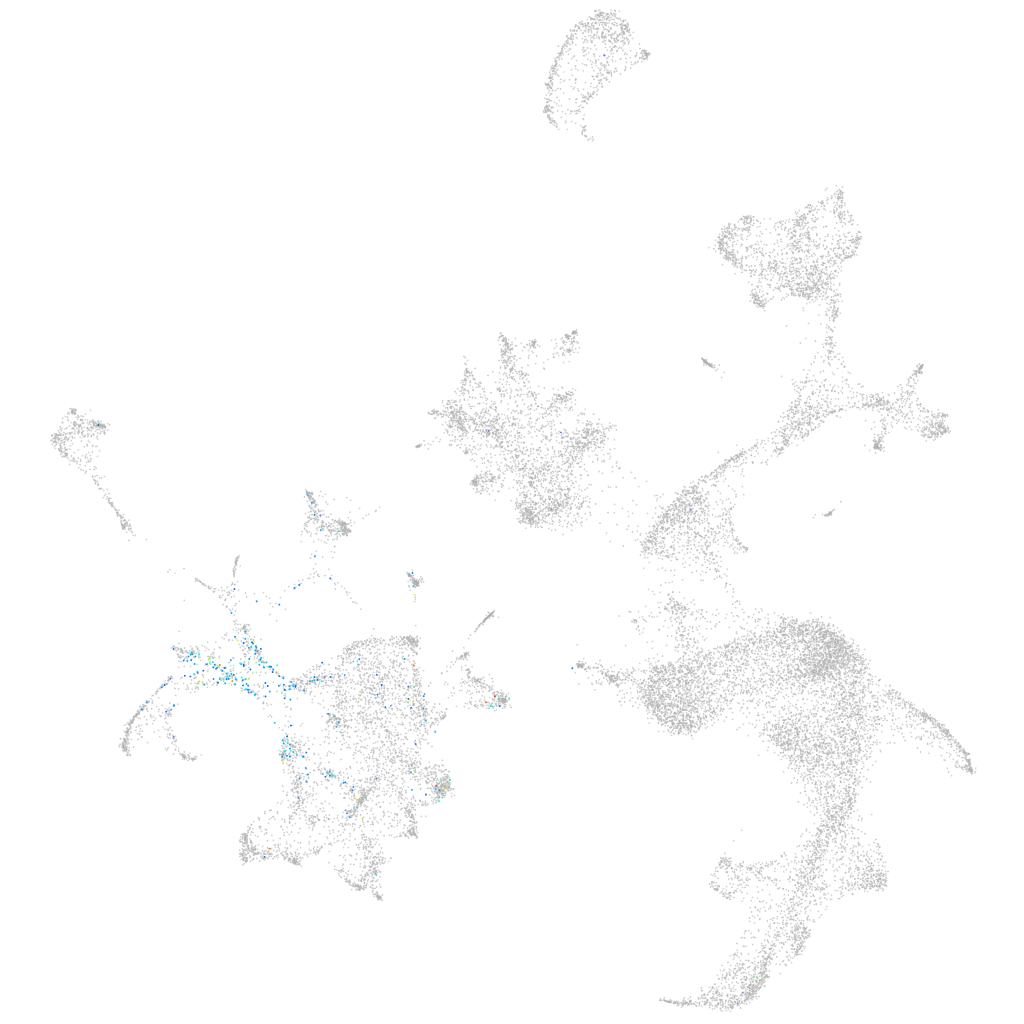
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 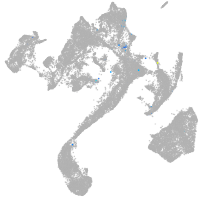 |
mural cells / non-skeletal muscle 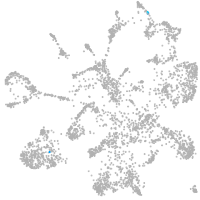 |
mesenchyme 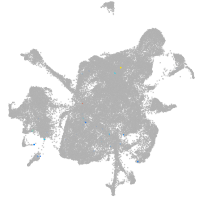 |
hematopoietic / vasculature 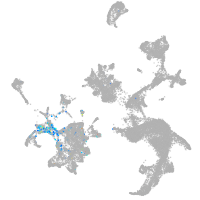 |
pronephros 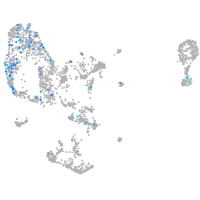 |
neural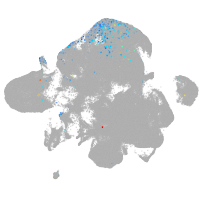 |
spinal cord / glia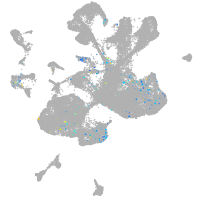 |
eye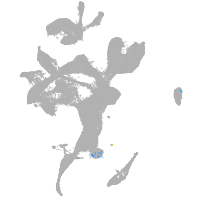 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| etv2 | 0.233 | prdx2 | -0.106 |
| si:dkey-52l18.4 | 0.210 | hbae1.1 | -0.103 |
| npas4l | 0.204 | hbae3 | -0.102 |
| egfl7 | 0.202 | hbbe1.3 | -0.100 |
| gbx2 | 0.185 | hbbe2 | -0.097 |
| tpm4a | 0.184 | hbbe1.1 | -0.095 |
| lmo2 | 0.182 | hbae1.3 | -0.095 |
| sox7 | 0.181 | cahz | -0.088 |
| tal1 | 0.180 | blvrb | -0.084 |
| fscn1a | 0.174 | hbbe1.2 | -0.082 |
| tmem88a | 0.168 | hemgn | -0.082 |
| ndnf | 0.167 | alas2 | -0.076 |
| dusp5 | 0.165 | tspo | -0.074 |
| colec12 | 0.165 | si:ch211-250g4.3 | -0.073 |
| wasf3b | 0.163 | slc4a1a | -0.072 |
| tjp1a | 0.160 | creg1 | -0.072 |
| si:ch73-364h19.1 | 0.157 | nt5c2l1 | -0.071 |
| BX469925.3 | 0.154 | gpx1a | -0.067 |
| hhex | 0.152 | fth1a | -0.067 |
| lin28a | 0.149 | epb41b | -0.067 |
| gata2a | 0.148 | zgc:163057 | -0.067 |
| flrt3 | 0.146 | si:ch211-207c6.2 | -0.066 |
| dact2 | 0.146 | tmem14ca | -0.061 |
| si:dkey-261h17.1 | 0.146 | nmt1b | -0.061 |
| ramp2 | 0.144 | tmod4 | -0.061 |
| flt4 | 0.143 | plac8l1 | -0.060 |
| sox18 | 0.143 | selenoj | -0.057 |
| marcksl1a | 0.141 | hbae5 | -0.053 |
| robo3 | 0.140 | si:ch211-227m13.1 | -0.050 |
| nxpe3 | 0.140 | si:dkey-240n22.3 | -0.049 |
| tspan7 | 0.140 | klf1 | -0.049 |
| akap12b | 0.139 | add2 | -0.049 |
| ildr2 | 0.139 | sptb | -0.048 |
| b3gnt7 | 0.139 | tfr1a | -0.048 |
| plk2b | 0.138 | hdr | -0.047 |