BUD23 rRNA methyltransferase and ribosome maturation factor
ZFIN 
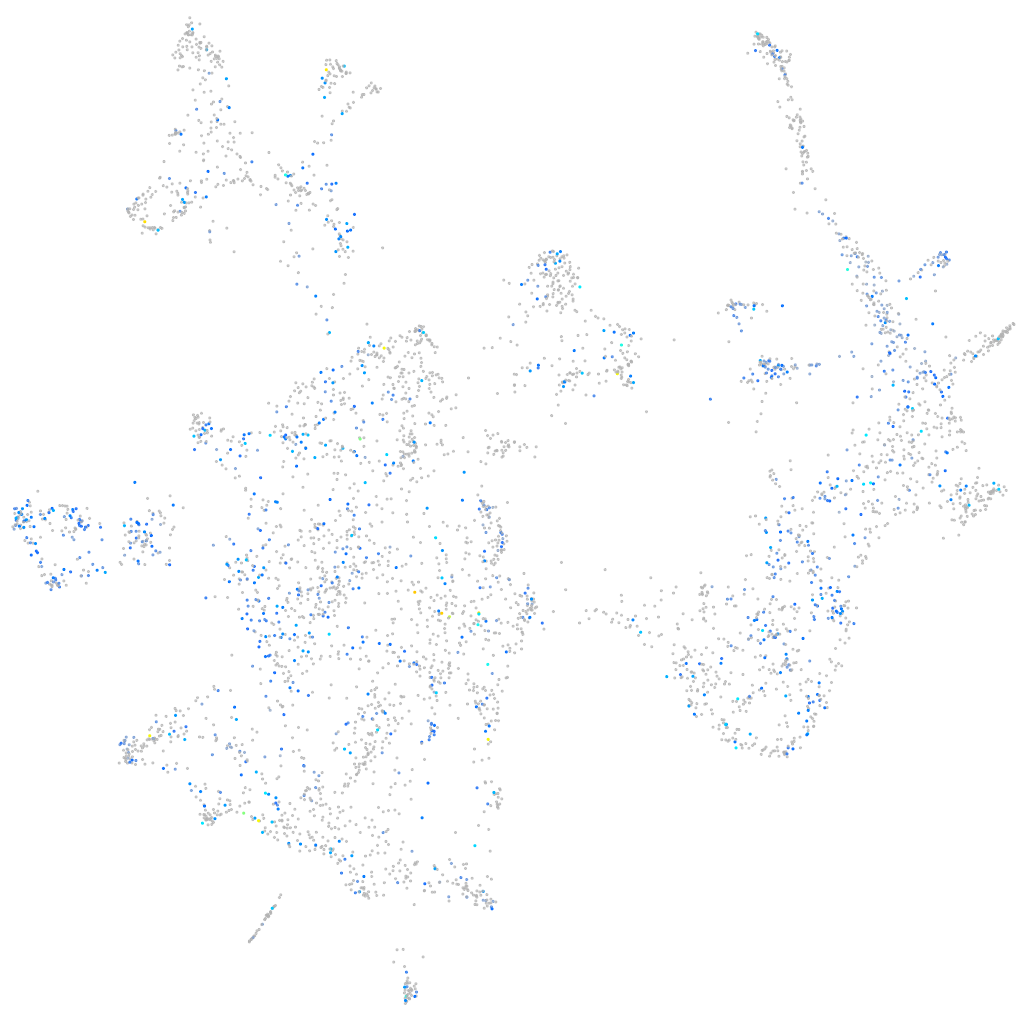
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nop10 | 0.219 | COX3 | -0.103 |
| npm1a | 0.215 | zgc:158463 | -0.095 |
| snu13b | 0.212 | mt-co2 | -0.095 |
| nop58 | 0.194 | CR383676.1 | -0.082 |
| fbl | 0.188 | pvalb1 | -0.082 |
| nop56 | 0.187 | kif1aa | -0.075 |
| dkc1 | 0.183 | tecpr1b | -0.075 |
| pa2g4a | 0.182 | pitpnab | -0.075 |
| rpl7l1 | 0.178 | NC-002333.17 | -0.075 |
| mcm7 | 0.177 | ptbp3 | -0.071 |
| nip7 | 0.176 | si:dkey-11c5.11 | -0.070 |
| tomm20a | 0.175 | tmprss3a | -0.068 |
| nhp2 | 0.173 | vsig10l2 | -0.068 |
| ncl | 0.170 | mt-co1 | -0.067 |
| si:ch211-217k17.7 | 0.167 | LOC110439372 | -0.067 |
| snrpd1 | 0.166 | osbpl1a | -0.066 |
| ranbp1 | 0.164 | tekt3 | -0.065 |
| snrpd2 | 0.164 | gpx2 | -0.065 |
| selenoh | 0.164 | calm1b | -0.065 |
| snrpf | 0.161 | tmbim1b | -0.064 |
| setb | 0.161 | BX005003.1 | -0.064 |
| mrto4 | 0.160 | h1f0 | -0.064 |
| snrpb | 0.159 | myo15ab | -0.063 |
| ddx39ab | 0.159 | eno1a | -0.063 |
| mak16 | 0.158 | cd164l2 | -0.063 |
| hspe1 | 0.158 | pvalb2 | -0.063 |
| c1qbp | 0.156 | atp2b1a | -0.063 |
| mcm6 | 0.156 | hsd3b7 | -0.063 |
| cnbpa | 0.156 | shisal1a | -0.063 |
| hmga1a | 0.156 | c16h7orf31 | -0.063 |
| cbx3a | 0.155 | emb | -0.063 |
| mcm4 | 0.155 | actc1b | -0.063 |
| ssb | 0.154 | chrna9 | -0.062 |
| wdr43 | 0.153 | CR925719.1 | -0.062 |
| hnrnpabb | 0.153 | atp1a3b | -0.062 |