"bloodthirsty-related gene family, member 1"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm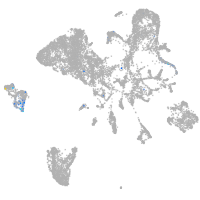 |
axial mesoderm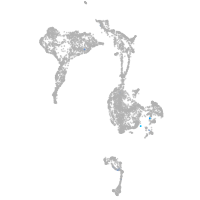 |
muscle 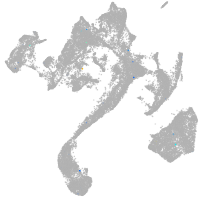 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 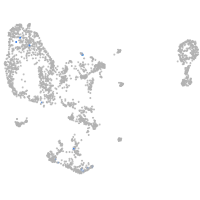 |
neural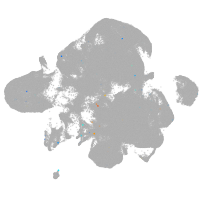 |
spinal cord / glia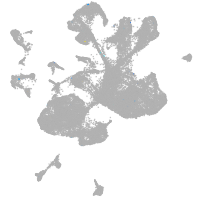 |
eye |
taste / olfactory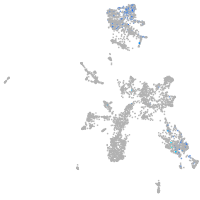 |
otic / lateral line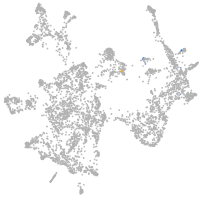 |
epidermis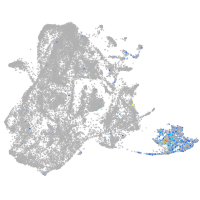 |
periderm |
fin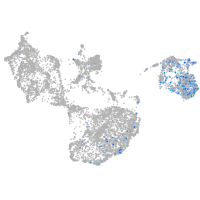 |
ionocytes / mucous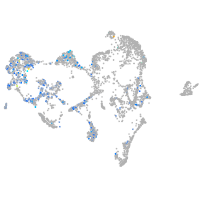 |
pigment cells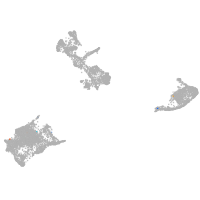 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkeyp-82a1.2 | 0.392 | rps5 | -0.043 |
| XLOC-018977 | 0.391 | rps26 | -0.042 |
| LOC108179274 | 0.257 | rpl22 | -0.038 |
| cplx3b | 0.250 | rps16 | -0.038 |
| fam129aa | 0.247 | rpl30 | -0.037 |
| fdxacb1 | 0.246 | naca | -0.033 |
| ftr14l | 0.235 | rpl36a | -0.033 |
| lim2.1 | 0.232 | calm2b | -0.028 |
| si:cabz01007807.1 | 0.222 | rpl18 | -0.027 |
| tnni2a.1 | 0.216 | jpt1b | -0.026 |
| ifit11 | 0.212 | h2afvb | -0.026 |
| LOC110438215 | 0.199 | cirbpb | -0.026 |
| si:ch211-114c17.1 | 0.191 | rps3 | -0.026 |
| slc37a4b | 0.183 | calm3b | -0.026 |
| XLOC-029303 | 0.183 | mt-co1 | -0.026 |
| olfcg6 | 0.177 | rps19 | -0.025 |
| slitrk3a | 0.176 | hnrnpa0l | -0.024 |
| gfra3 | 0.175 | eif3f | -0.024 |
| dbnla | 0.166 | hmgn7 | -0.024 |
| si:dkey-242h9.3 | 0.164 | ppp1cbl | -0.024 |
| pcdh8 | 0.163 | romo1 | -0.023 |
| si:ch211-161c3.5 | 0.162 | rpsa | -0.023 |
| zgc:112964 | 0.160 | hnrnpaba | -0.023 |
| mat2al | 0.157 | eef1b2 | -0.023 |
| lgals17 | 0.155 | rpl10 | -0.023 |
| trim63b | 0.152 | ndufb7 | -0.023 |
| tagln | 0.147 | bsg | -0.023 |
| hbae5 | 0.147 | cox6c | -0.022 |
| si:ch211-43f4.1 | 0.145 | rplp2l | -0.022 |
| RASSF5 | 0.142 | calm1a | -0.022 |
| eif4g3a | 0.139 | wbp2nl | -0.022 |
| fbp2 | 0.139 | tubb2b | -0.022 |
| chodl | 0.136 | eno3 | -0.022 |
| nectin4a | 0.136 | rab34b | -0.022 |
| ston2 | 0.131 | nme2b.1 | -0.022 |




