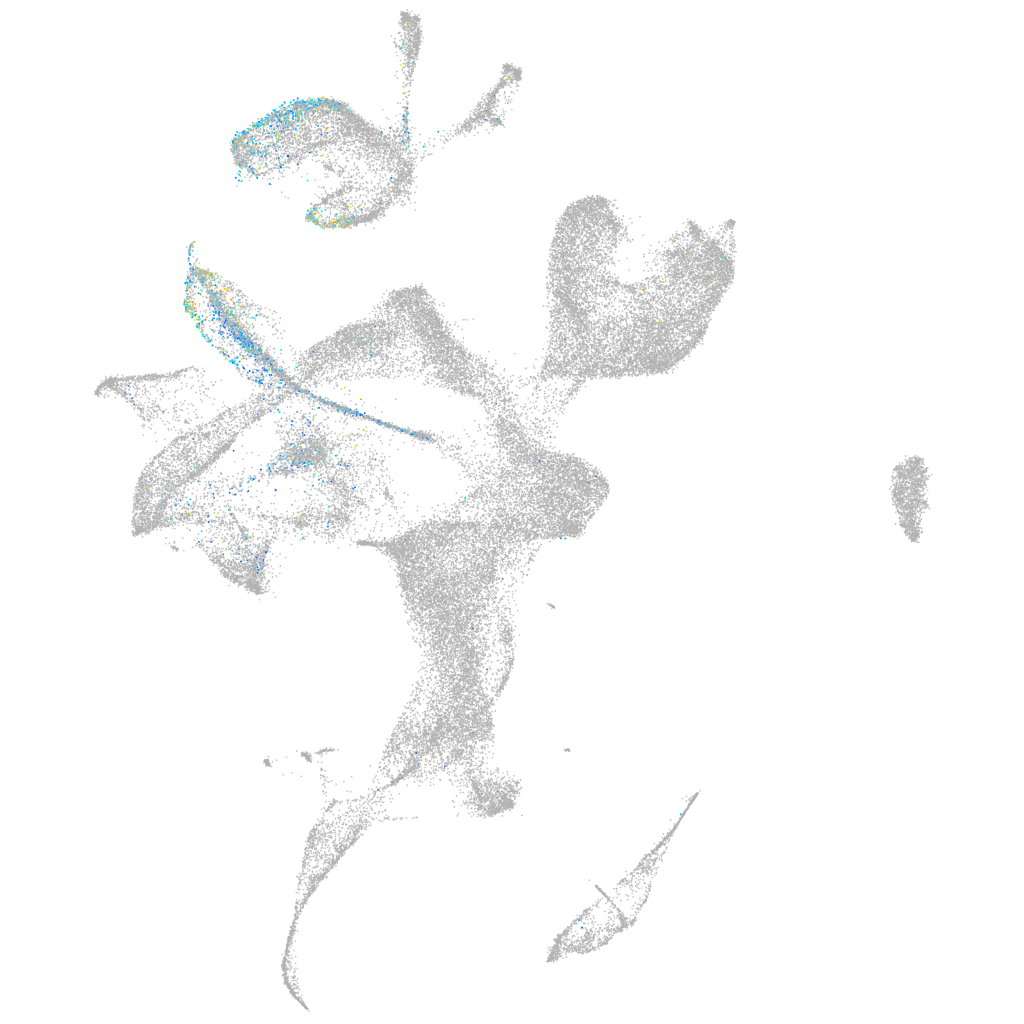BTB (POZ) domain containing 3b
ZFIN 
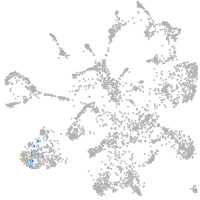
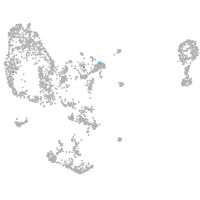
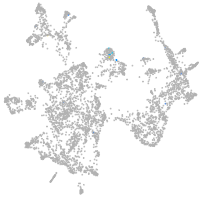
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.226 | hmgb2a | -0.165 |
| eno2 | 0.222 | pcna | -0.084 |
| rbpms2a | 0.217 | stmn1a | -0.083 |
| stx1b | 0.214 | mdka | -0.083 |
| stxbp1a | 0.213 | fabp7a | -0.079 |
| sv2a | 0.212 | rrm1 | -0.078 |
| rtn1b | 0.204 | dut | -0.078 |
| stmn2a | 0.200 | hmga1a | -0.077 |
| rbpms2b | 0.200 | msi1 | -0.077 |
| si:ch211-129p13.1 | 0.197 | nutf2l | -0.076 |
| zgc:153426 | 0.197 | lbr | -0.076 |
| ywhag2 | 0.197 | mki67 | -0.074 |
| syt1a | 0.195 | ccnd1 | -0.074 |
| sncb | 0.193 | msna | -0.073 |
| zgc:65894 | 0.192 | ahcy | -0.073 |
| elavl4 | 0.191 | chaf1a | -0.073 |
| cplx2l | 0.190 | tuba8l | -0.072 |
| snap25a | 0.190 | ccna2 | -0.071 |
| islr2 | 0.188 | mcm7 | -0.070 |
| tfap2e | 0.186 | neurod4 | -0.070 |
| tkta | 0.185 | selenoh | -0.070 |
| ppp1r14ba | 0.184 | hmgb2b | -0.067 |
| tuba2 | 0.184 | her15.1 | -0.067 |
| irx4a | 0.181 | rpa3 | -0.067 |
| gnao1a | 0.177 | tuba8l4 | -0.066 |
| gng3 | 0.176 | cks1b | -0.066 |
| stmn1b | 0.176 | banf1 | -0.066 |
| ebf3a | 0.175 | dek | -0.066 |
| celf5a | 0.175 | fen1 | -0.065 |
| sprn | 0.175 | crx | -0.065 |
| grin1a | 0.174 | lig1 | -0.065 |
| xpr1a | 0.173 | rpa2 | -0.064 |
| inab | 0.172 | id1 | -0.064 |
| vsnl1b | 0.171 | mibp | -0.064 |
| mllt11 | 0.169 | rrm2 | -0.064 |