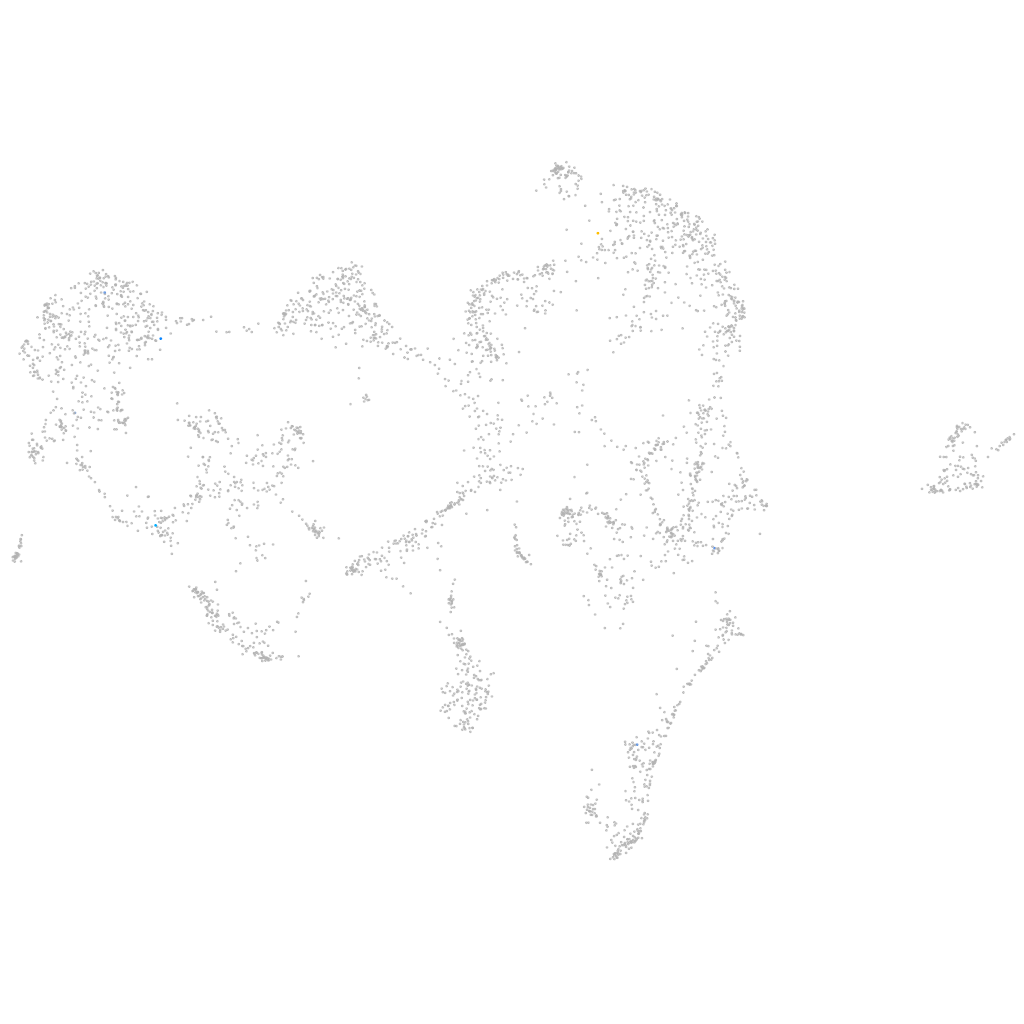
barttin CLCNK-type chloride channel accessory beta subunit
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 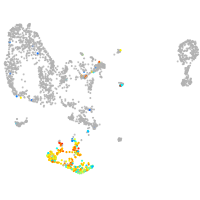 |
neural |
spinal cord / glia |
eye |
taste / olfactory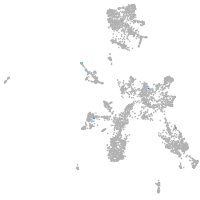 |
otic / lateral line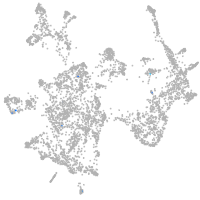 |
epidermis |
periderm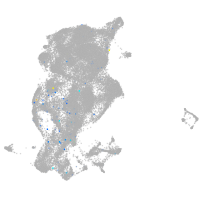 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chrnd | 0.669 | rps26 | -0.058 |
| CR376766.2 | 0.617 | rps3 | -0.045 |
| slitrk5a | 0.491 | rpl23 | -0.040 |
| prdm8b | 0.463 | eef1b2 | -0.039 |
| bdh2 | 0.456 | cox8a | -0.038 |
| si:dkey-234l24.1 | 0.388 | rpl22l1 | -0.037 |
| SMARCA4 | 0.388 | si:ch1073-429i10.3.1 | -0.035 |
| si:dkey-29h14.10 | 0.379 | eif3ea | -0.033 |
| npffr1l1 | 0.373 | atp5mc3a | -0.033 |
| mpeg1.2 | 0.305 | rpl32 | -0.032 |
| tjap1 | 0.271 | cirbpb | -0.032 |
| LOC100005864 | 0.271 | eef2b | -0.032 |
| wfs1b | 0.269 | atp5pf | -0.032 |
| znf526 | 0.267 | tegt | -0.031 |
| ATE1 | 0.261 | atp5f1e | -0.031 |
| clstn3 | 0.260 | ap2m1a | -0.031 |
| tapbp.1 | 0.258 | COX5B | -0.031 |
| phykpl | 0.242 | rps13 | -0.031 |
| LOC103909784 | 0.238 | cox6a1 | -0.031 |
| cryba2b | 0.231 | ssr2 | -0.030 |
| hoxa10b | 0.221 | tmem258 | -0.030 |
| pcloa | 0.207 | hmgn6 | -0.030 |
| znf574 | 0.201 | cldnh | -0.030 |
| si:dkey-217d24.6 | 0.200 | brk1 | -0.029 |
| BX284696.1 | 0.198 | fthl27 | -0.029 |
| rubcn | 0.194 | atp5pd | -0.029 |
| PEBP4 | 0.191 | arf5 | -0.029 |
| si:dkey-73p2.3 | 0.188 | rnaseka | -0.029 |
| plekhm1 | 0.186 | cox7c | -0.029 |
| bcl2l11 | 0.184 | rab10 | -0.029 |
| eefsec | 0.184 | atp5if1b | -0.028 |
| edil3a | 0.183 | ndufa4l | -0.028 |
| s100t | 0.181 | atp5fa1 | -0.028 |
| BX005012.4 | 0.180 | sf3b6 | -0.028 |
| fam151b | 0.180 | uqcrq | -0.028 |