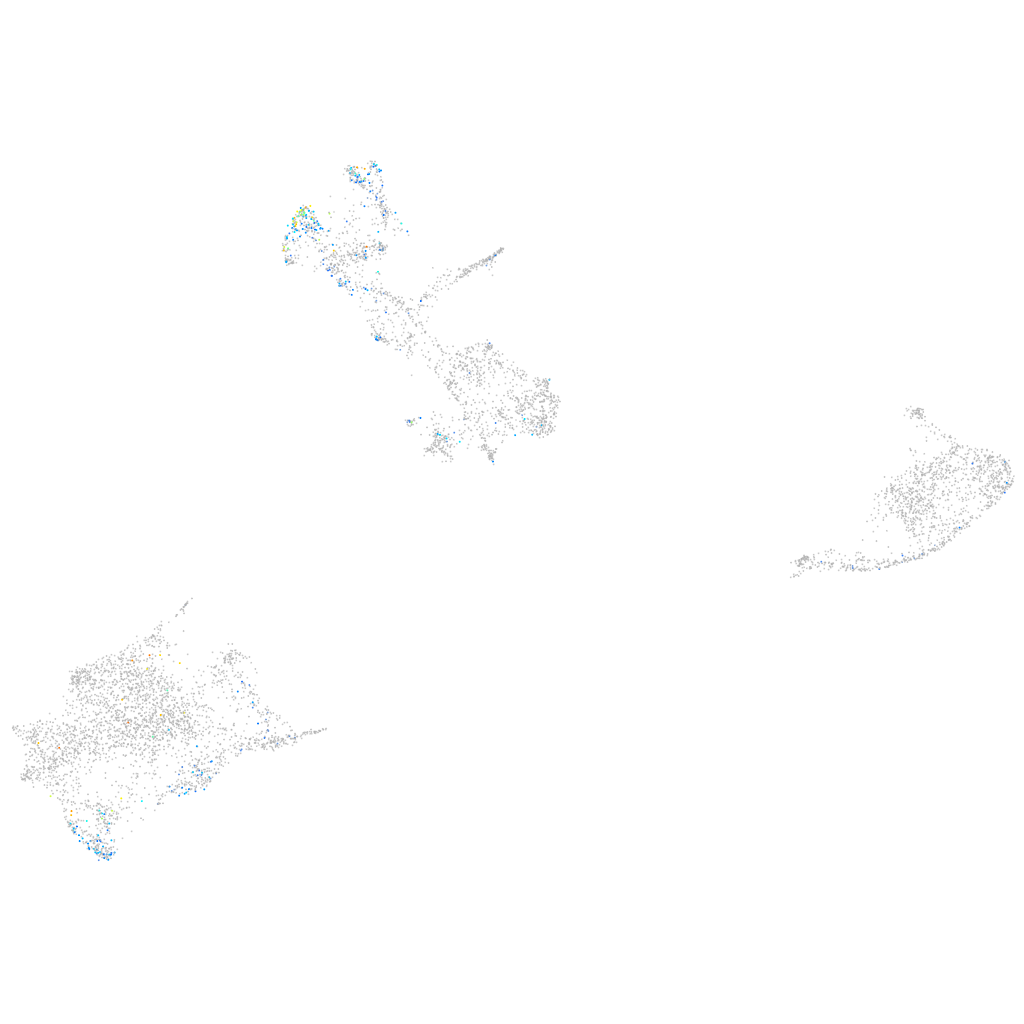
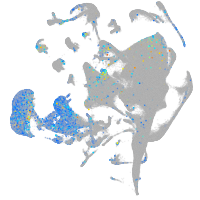
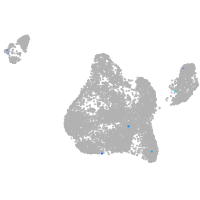
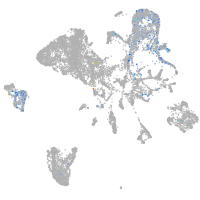
BCL2 family apoptosis regulator BOK a
ZFIN 
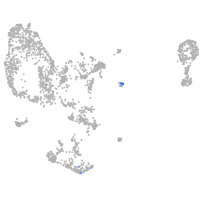
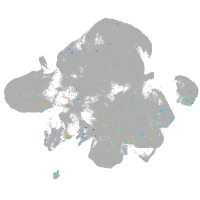
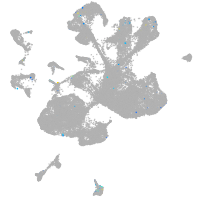
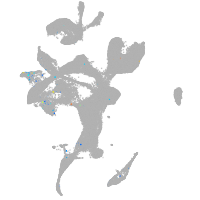
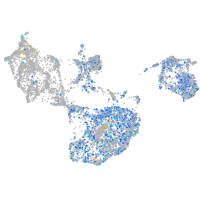
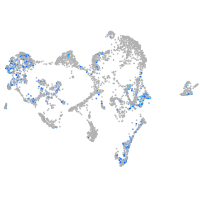
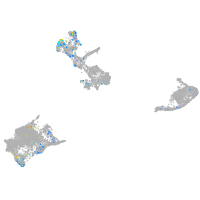
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| csf1a | 0.275 | gch2 | -0.092 |
| ppp1r1c | 0.262 | qdpra | -0.086 |
| sytl2b | 0.260 | cotl1 | -0.085 |
| akap12a | 0.257 | hbbe1.3 | -0.085 |
| syne1b | 0.243 | tyrp1b | -0.083 |
| fhl3a | 0.237 | pmela | -0.082 |
| lypc | 0.235 | pts | -0.081 |
| prx | 0.233 | zgc:91968 | -0.080 |
| ponzr1 | 0.229 | akr1b1 | -0.080 |
| CT737162.3 | 0.221 | marcksl1b | -0.077 |
| tpd52l1 | 0.220 | tyrp1a | -0.075 |
| sdc2 | 0.218 | pcbd1 | -0.074 |
| sytl2a | 0.215 | fabp3 | -0.074 |
| si:ch211-256m1.8 | 0.213 | dct | -0.071 |
| ppfibp1a | 0.212 | gstt1a | -0.070 |
| si:dkey-197i20.6 | 0.212 | anxa1a | -0.070 |
| plecb | 0.210 | oca2 | -0.069 |
| gpnmb | 0.210 | si:dkey-151g10.6 | -0.068 |
| cmtm6 | 0.209 | tyr | -0.065 |
| tcirg1a | 0.208 | pah | -0.065 |
| alx4a | 0.207 | vat1 | -0.063 |
| gbx2 | 0.207 | si:ch211-222l21.1 | -0.062 |
| fhl2a | 0.206 | tubb2b | -0.060 |
| apoda.1 | 0.205 | prkar1b | -0.060 |
| alx4b | 0.200 | ckbb | -0.060 |
| itga6b | 0.200 | marcksl1a | -0.060 |
| efhd2 | 0.199 | slc24a5 | -0.060 |
| kank3 | 0.197 | tuba1c | -0.059 |
| ltk | 0.197 | marcksb | -0.059 |
| dennd2db | 0.196 | mtbl | -0.058 |
| si:ch73-89b15.3 | 0.196 | kita | -0.058 |
| pltp | 0.195 | hbae1.1 | -0.057 |
| tmeff2a | 0.191 | hbae3 | -0.057 |
| myo5b | 0.186 | tspan10 | -0.056 |
| cdh11 | 0.185 | tmem243a | -0.056 |