bone morphogenetic protein 4
ZFIN 
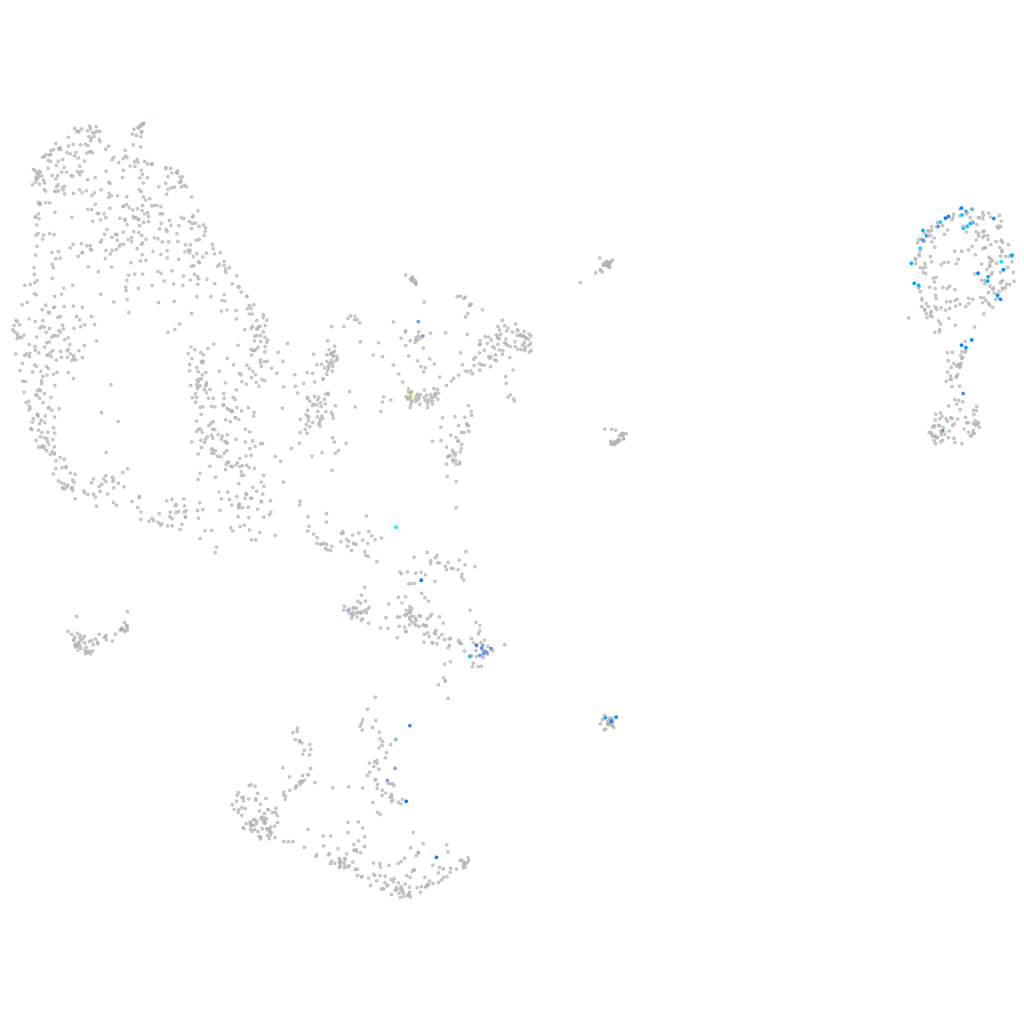
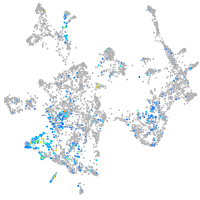
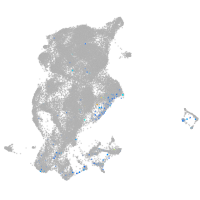
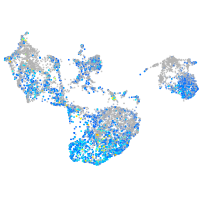
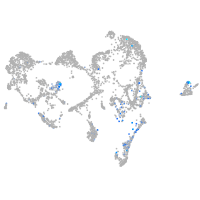
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ved | 0.372 | rpl37 | -0.219 |
| szl | 0.344 | atp5f1b | -0.211 |
| cdx1a | 0.320 | rps10 | -0.211 |
| tbx16 | 0.318 | atp5fa1 | -0.204 |
| bambia | 0.302 | rps17 | -0.199 |
| apoc1 | 0.292 | atp1b1a | -0.199 |
| CABZ01044023.1 | 0.279 | zgc:114188 | -0.197 |
| apoeb | 0.278 | suclg1 | -0.183 |
| vent | 0.273 | atp5mc1 | -0.182 |
| vox | 0.272 | atp5meb | -0.181 |
| trim101 | 0.269 | zgc:56493 | -0.178 |
| cdx4 | 0.266 | romo1 | -0.178 |
| cx43.4 | 0.265 | slc25a5 | -0.176 |
| XLOC-016197 | 0.262 | atp5f1c | -0.174 |
| tmem266 | 0.259 | atp1a1a.4 | -0.173 |
| lig1 | 0.259 | atp5l | -0.172 |
| CT030712.1 | 0.242 | atp5mc3b | -0.170 |
| hoxa9b | 0.241 | eno3 | -0.170 |
| CT573139.2 | 0.232 | cox8a | -0.170 |
| hspb1 | 0.229 | si:ch211-139a5.9 | -0.170 |
| LOC101884310 | 0.228 | mt-nd1 | -0.169 |
| nop58 | 0.228 | idh2 | -0.169 |
| ripk2 | 0.228 | cox5aa | -0.168 |
| zgc:66024 | 0.225 | tpi1b | -0.163 |
| wnt5b | 0.223 | vdac2 | -0.161 |
| akap12b | 0.221 | cox7a2a | -0.160 |
| snrnp70 | 0.220 | ndrg1a | -0.159 |
| ncl | 0.219 | cox6a1 | -0.159 |
| asph | 0.219 | atp5pb | -0.159 |
| gpr35.1 | 0.219 | atp5if1b | -0.159 |
| hnrnpa1a | 0.218 | mt-atp6 | -0.158 |
| rab25a | 0.218 | vdac3 | -0.157 |
| anp32e | 0.218 | COX5B | -0.157 |
| nop2 | 0.218 | ezra | -0.157 |
| nucks1a | 0.216 | ldhba | -0.156 |