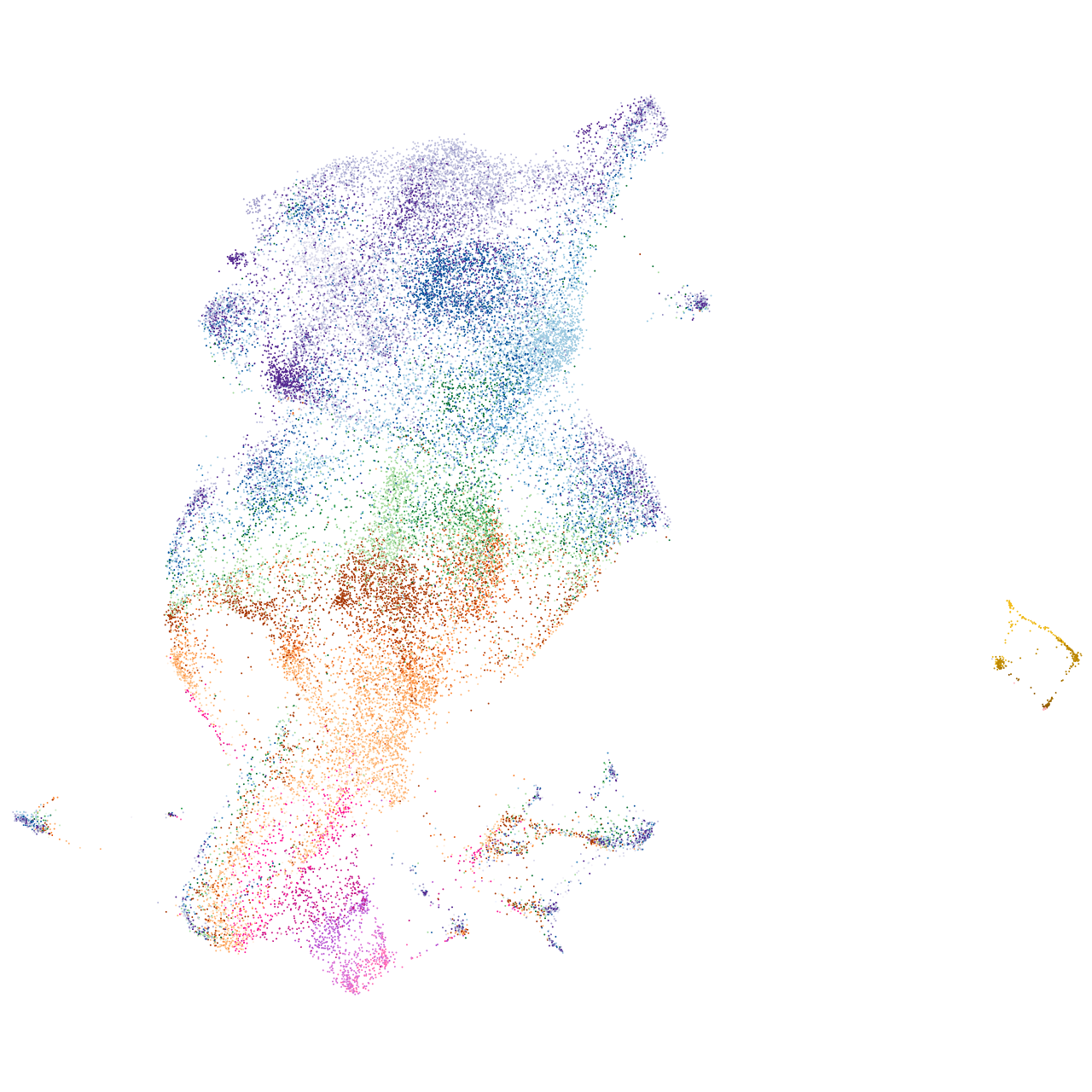"biogenesis of lysosomal organelles complex-1, subunit 3"
ZFIN 
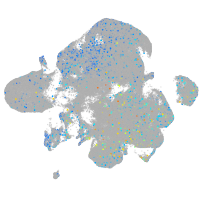
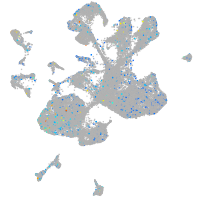
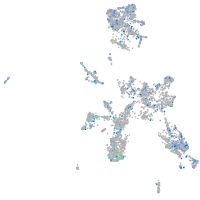
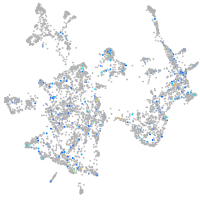
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ran | 0.085 | CABZ01072614.1 | -0.046 |
| si:ch73-52f15.5 | 0.084 | tmsb4x | -0.046 |
| glo1 | 0.081 | alox12 | -0.035 |
| eef1g | 0.079 | LOC110439372 | -0.034 |
| fuca1.1 | 0.079 | icn | -0.033 |
| eef1b2 | 0.076 | XLOC-041945 | -0.032 |
| rsl24d1 | 0.075 | hbbe1.3 | -0.031 |
| tlcd1 | 0.074 | si:cabz01007794.1 | -0.030 |
| gabarapb | 0.074 | zgc:158463 | -0.030 |
| cox6b2 | 0.073 | hbae3 | -0.030 |
| atp5mc3b | 0.072 | CT027638.1 | -0.027 |
| cox4i1 | 0.072 | cldn17 | -0.026 |
| romo1 | 0.072 | krt92 | -0.024 |
| cox7a2l | 0.072 | NC-002333.17 | -0.024 |
| npc2 | 0.071 | LOC110439627 | -0.024 |
| atox1 | 0.071 | myhz1.1 | -0.023 |
| eef1da | 0.071 | evplb | -0.023 |
| eif4ebp1 | 0.070 | ppl | -0.023 |
| si:dkey-112e7.2 | 0.070 | mt-atp8 | -0.023 |
| prdx5 | 0.070 | slx1b | -0.023 |
| vdac2 | 0.069 | NC-002333.4 | -0.023 |
| ctsla | 0.069 | ckma | -0.022 |
| prdx2 | 0.069 | ttn.1 | -0.022 |
| pycard | 0.069 | cyp2y3 | -0.022 |
| adi1 | 0.068 | ost4 | -0.022 |
| ybx1 | 0.068 | AL935186.9 | -0.021 |
| slc25a3b | 0.068 | actc1b | -0.021 |
| eif3f | 0.067 | actb1 | -0.021 |
| znf706 | 0.067 | pvalb2 | -0.021 |
| gtpbp4 | 0.066 | krt18a.1 | -0.021 |
| nme2b.1 | 0.066 | hbae1.1 | -0.021 |
| chmp5b | 0.066 | duox | -0.020 |
| fxyd6l | 0.066 | pvalb1 | -0.020 |
| map1lc3a | 0.065 | hbbe1.2 | -0.020 |
| rpl19 | 0.065 | DDX17 | -0.020 |