"biogenesis of lysosomal organelles complex-1, subunit 3"
ZFIN 
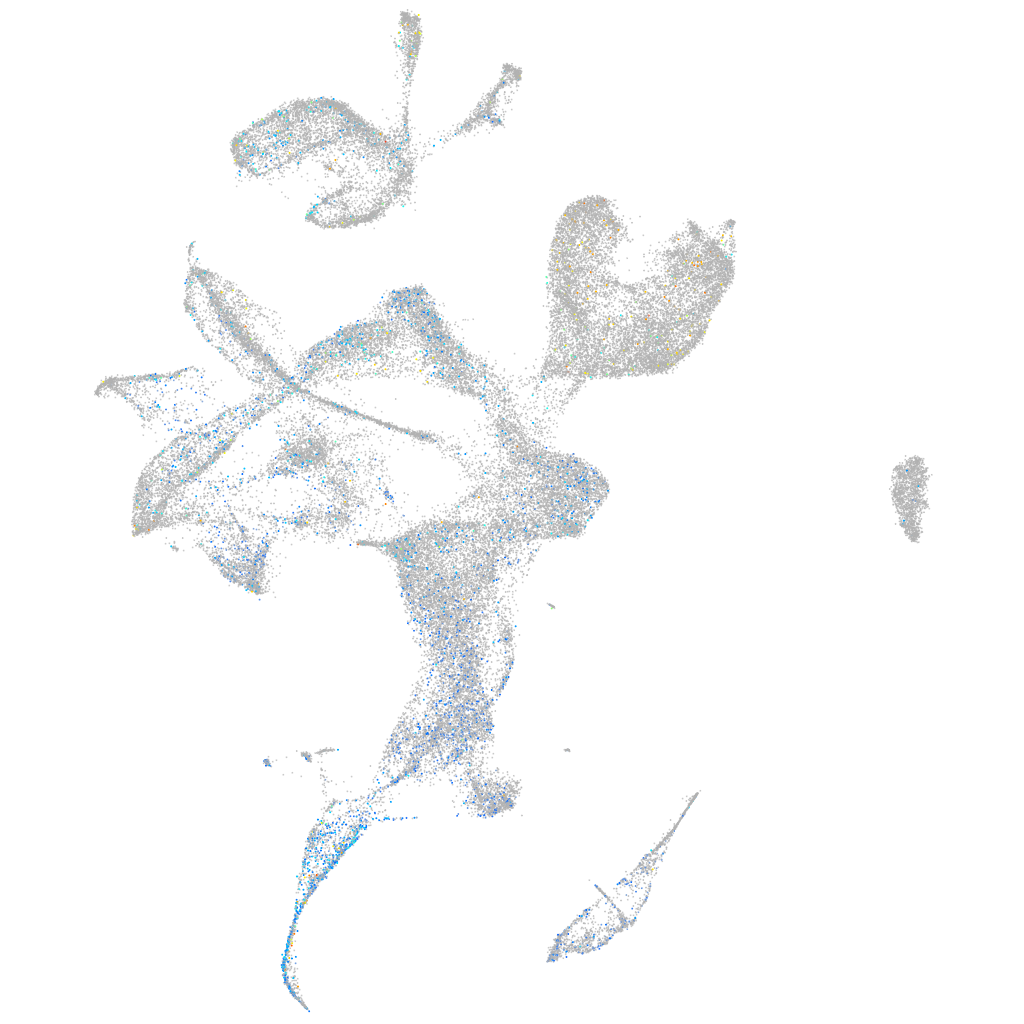
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr143 | 0.180 | ptmab | -0.083 |
| dct | 0.179 | si:ch73-1a9.3 | -0.076 |
| slc45a2 | 0.178 | ptmaa | -0.051 |
| pmela | 0.169 | hmgb1a | -0.048 |
| tspan36 | 0.169 | gpm6aa | -0.047 |
| tyrp1b | 0.169 | nova2 | -0.047 |
| pmelb | 0.167 | si:ch211-222l21.1 | -0.045 |
| mitfa | 0.159 | hmgn6 | -0.042 |
| tyrp1a | 0.158 | scrt2 | -0.041 |
| tspan10 | 0.158 | mdkb | -0.040 |
| rab38 | 0.155 | bcl2l10 | -0.038 |
| oca2 | 0.153 | marcksl1b | -0.038 |
| cracr2ab | 0.153 | marcksb | -0.038 |
| bace2 | 0.152 | tp53inp2 | -0.037 |
| slc24a5 | 0.150 | si:ch211-137a8.4 | -0.036 |
| qdpra | 0.150 | tuba1a | -0.036 |
| fabp11b | 0.150 | marcksl1a | -0.034 |
| rab32a | 0.148 | hmgb1b | -0.033 |
| msnb | 0.146 | rorb | -0.033 |
| triobpa | 0.146 | rtn1a | -0.032 |
| pttg1ipb | 0.142 | hmgb3a | -0.031 |
| gstp1 | 0.142 | neurod4 | -0.031 |
| agtrap | 0.142 | vsx1 | -0.029 |
| tyr | 0.142 | hnrnpaba | -0.029 |
| mb | 0.141 | hmgn2 | -0.029 |
| rgrb | 0.140 | chd4a | -0.029 |
| zgc:110591 | 0.140 | myt1b | -0.029 |
| FP085398.1 | 0.136 | zfhx4 | -0.029 |
| stra6 | 0.134 | FO082781.1 | -0.027 |
| pcbd1 | 0.132 | cirbpb | -0.027 |
| trpm1b | 0.130 | hunk | -0.027 |
| fgfbp1a | 0.130 | rcc2 | -0.027 |
| tm6sf2 | 0.129 | epb41a | -0.027 |
| rlbp1b | 0.127 | sox11b | -0.026 |
| si:ch73-86n18.1 | 0.127 | CR383676.1 | -0.026 |