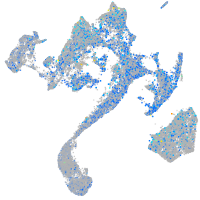
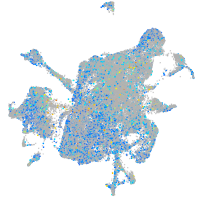
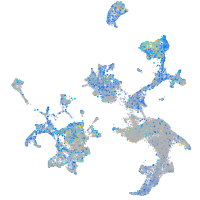
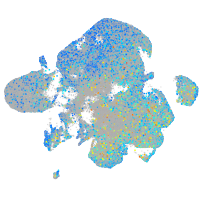
"biogenesis of lysosomal organelles complex-1, subunit 2"
ZFIN 
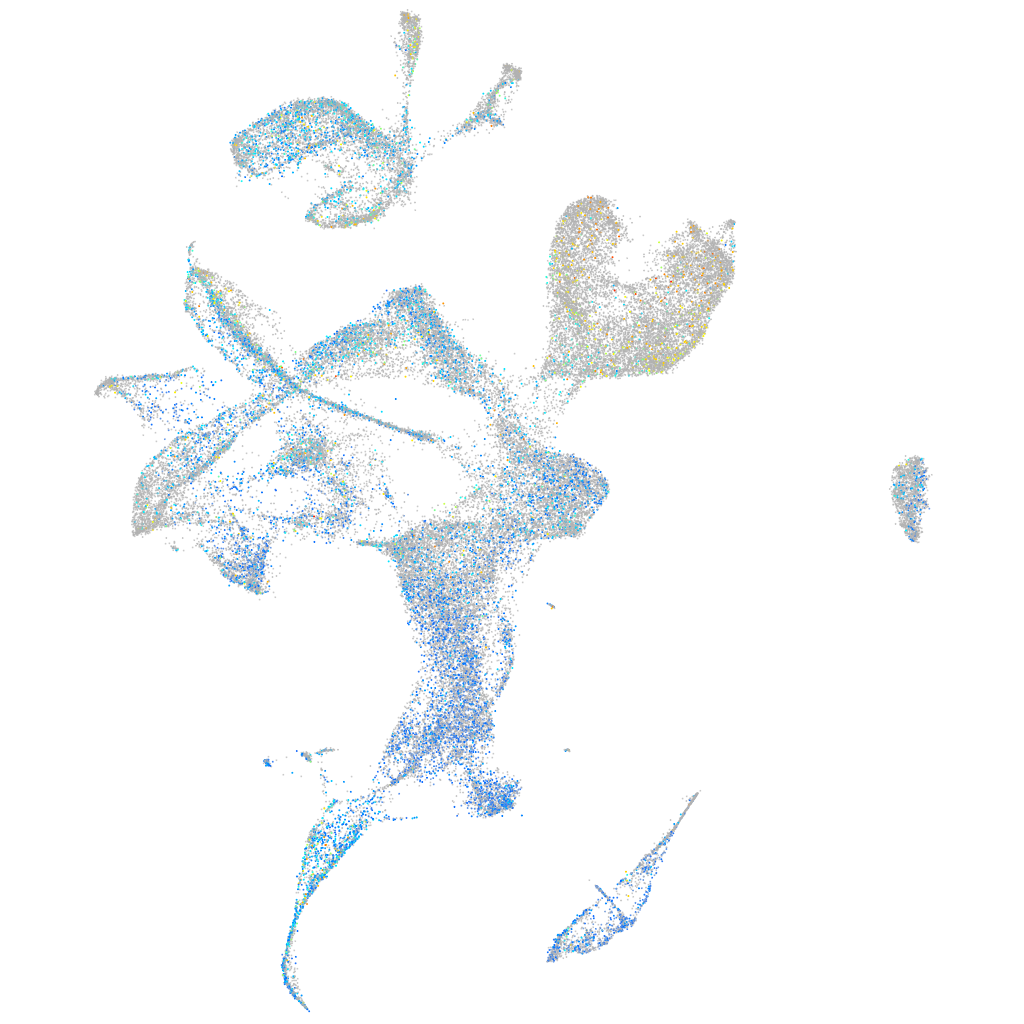
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr143 | 0.135 | vsx1 | -0.067 |
| slc45a2 | 0.134 | si:ch73-1a9.3 | -0.060 |
| dct | 0.134 | neurod4 | -0.057 |
| tspan36 | 0.134 | histh1l | -0.056 |
| rab38 | 0.127 | ptmab | -0.055 |
| tyrp1b | 0.127 | syt5b | -0.053 |
| pmela | 0.125 | gnb3a | -0.053 |
| pmelb | 0.124 | samsn1a | -0.047 |
| tyrp1a | 0.121 | epb41a | -0.046 |
| qdpra | 0.120 | si:ch1073-83n3.2 | -0.045 |
| agtrap | 0.118 | cplx4a | -0.045 |
| tspan10 | 0.118 | nrn1lb | -0.044 |
| cracr2ab | 0.117 | calb2b | -0.042 |
| rab32a | 0.115 | si:ch73-256g18.2 | -0.041 |
| zgc:110591 | 0.115 | mdkb | -0.040 |
| mitfa | 0.114 | vamp1 | -0.039 |
| pcbd1 | 0.114 | hunk | -0.038 |
| gstp1 | 0.112 | stx3a | -0.038 |
| fabp11b | 0.111 | CR383676.1 | -0.038 |
| slc24a5 | 0.111 | RIC3 | -0.037 |
| bace2 | 0.111 | pcp4l1 | -0.037 |
| oca2 | 0.108 | hes2.2 | -0.037 |
| triobpa | 0.108 | ppp1r1b | -0.036 |
| pttg1ipb | 0.108 | npdc1a | -0.036 |
| tyr | 0.107 | cabp2a | -0.036 |
| FP085398.1 | 0.106 | BX663503.1 | -0.035 |
| prdx2 | 0.105 | btg1 | -0.035 |
| si:dkey-21a6.5 | 0.104 | LOC108180196 | -0.034 |
| vat1 | 0.104 | gng13b | -0.034 |
| msnb | 0.104 | tp53inp1 | -0.034 |
| rnaseka | 0.103 | zgc:153395 | -0.034 |
| col4a5 | 0.102 | zgc:77784 | -0.033 |
| si:ch73-86n18.1 | 0.100 | ndrg1b | -0.033 |
| sparc | 0.099 | pvalb2 | -0.033 |
| sytl2b | 0.099 | anp32e | -0.033 |