BCL6B transcription repressor
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 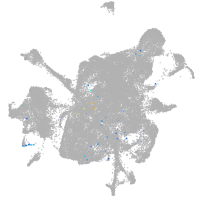 |
hematopoietic / vasculature  |
pronephros 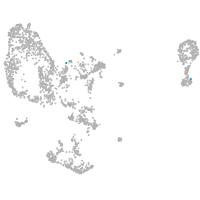 |
neural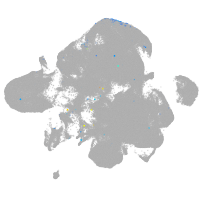 |
spinal cord / glia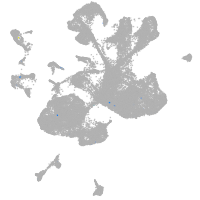 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101882826 | 0.201 | gpm6aa | -0.027 |
| tie1 | 0.178 | tuba1c | -0.026 |
| erg | 0.175 | elavl3 | -0.026 |
| hbl2 | 0.169 | rtn1a | -0.025 |
| pgfb | 0.159 | stmn1b | -0.025 |
| notchl | 0.150 | hmgb1b | -0.023 |
| scarf1 | 0.122 | tmsb | -0.022 |
| myct1a | 0.110 | ckbb | -0.020 |
| rasip1 | 0.106 | hmgb3a | -0.020 |
| CT573423.1 | 0.106 | si:dkey-276j7.1 | -0.020 |
| cdh5 | 0.104 | zc4h2 | -0.020 |
| egfl7 | 0.103 | hmgb1a | -0.019 |
| slc9a3r1b | 0.102 | gng3 | -0.019 |
| BX119963.4 | 0.096 | sncb | -0.019 |
| arhgap27l | 0.095 | gpm6ab | -0.019 |
| stab1 | 0.093 | nova1 | -0.018 |
| pecam1 | 0.091 | si:dkeyp-75h12.5 | -0.018 |
| kdrl | 0.090 | elavl4 | -0.018 |
| timp2b | 0.089 | nova2 | -0.017 |
| LOC101884756 | 0.089 | rtn1b | -0.017 |
| scpp8 | 0.088 | myt1a | -0.017 |
| ecscr | 0.087 | id4 | -0.017 |
| si:ch211-113e8.9 | 0.085 | rnasekb | -0.017 |
| si:ch211-241f5.3 | 0.084 | gap43 | -0.016 |
| si:ch73-334d15.2 | 0.082 | stmn2a | -0.016 |
| afap1l1b | 0.082 | fez1 | -0.016 |
| XLOC-039757 | 0.081 | myt1b | -0.016 |
| egr2b | 0.080 | cspg5a | -0.016 |
| tek | 0.076 | ptmab | -0.016 |
| fam83c | 0.073 | ptmaa | -0.015 |
| stim1b | 0.072 | CU467822.1 | -0.015 |
| frem1b | 0.072 | atp6v0cb | -0.015 |
| exoc3l2a | 0.071 | tuba1a | -0.015 |
| XLOC-014079 | 0.071 | zfhx3 | -0.015 |
| sat1a.1 | 0.071 | fam168a | -0.015 |