BBX high mobility group box domain containing
ZFIN 
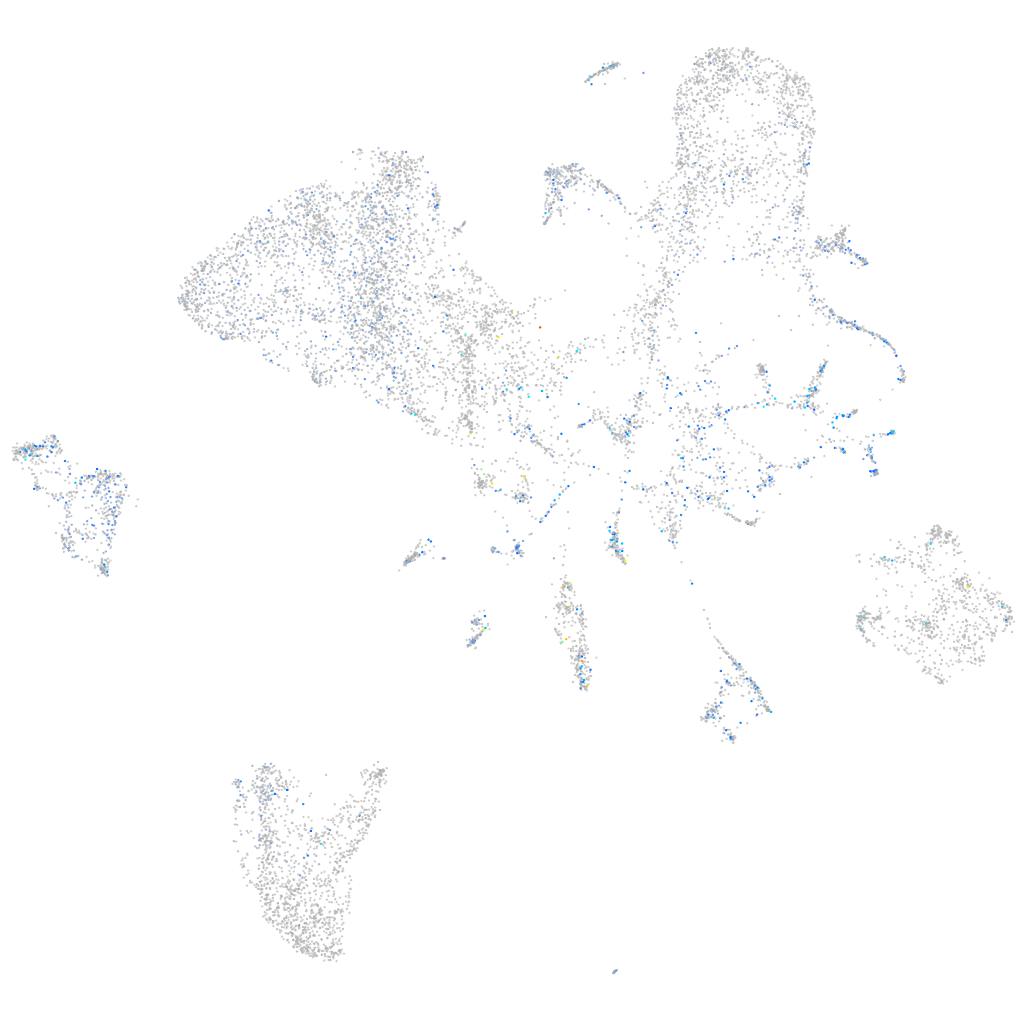
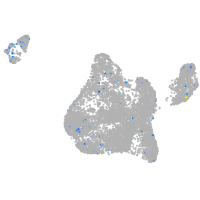
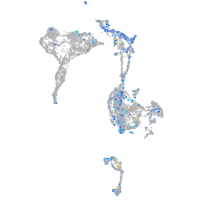
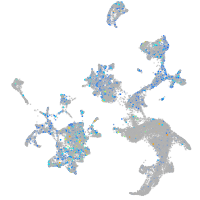
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.138 | gapdh | -0.088 |
| anxa4 | 0.130 | gamt | -0.086 |
| ier2b | 0.127 | gatm | -0.081 |
| LOC101884310 | 0.125 | zgc:136461 | -0.079 |
| calm1a | 0.125 | CELA1 (1 of many) | -0.079 |
| cldn7b | 0.124 | spink4 | -0.078 |
| elovl1b | 0.122 | prss59.1 | -0.078 |
| pax6b | 0.122 | ela2 | -0.078 |
| ccni | 0.122 | ela2l | -0.077 |
| epcam | 0.122 | cpb1 | -0.076 |
| si:ch1073-429i10.3.1 | 0.122 | cpa5 | -0.076 |
| oaz1b | 0.121 | cpa4 | -0.075 |
| tsc22d1 | 0.121 | amy2a | -0.075 |
| jun | 0.120 | sycn.2 | -0.075 |
| atf4b | 0.119 | c6ast4 | -0.075 |
| tmem59 | 0.118 | ela3l | -0.075 |
| scg3 | 0.115 | nupr1b | -0.075 |
| si:dkey-112e7.2 | 0.114 | zgc:112160 | -0.075 |
| spint2 | 0.113 | ctrb1 | -0.074 |
| capns1b | 0.112 | cel.2 | -0.074 |
| calm1b | 0.112 | cel.1 | -0.074 |
| ubc | 0.112 | prss1 | -0.073 |
| litaf | 0.112 | zgc:92137 | -0.073 |
| gpc1a | 0.112 | fep15 | -0.073 |
| gnb1a | 0.112 | prss59.2 | -0.073 |
| kdm6bb | 0.111 | cpa1 | -0.073 |
| btg2 | 0.111 | ahcy | -0.072 |
| pfn1 | 0.111 | endou | -0.071 |
| bnip3lb | 0.111 | si:ch211-240l19.5 | -0.070 |
| rnasekb | 0.110 | erp27 | -0.070 |
| zgc:101731 | 0.110 | mat1a | -0.069 |
| si:ch211-239f4.1 | 0.110 | apoda.2 | -0.069 |
| cldnb | 0.110 | apoc2 | -0.069 |
| ptbp3 | 0.110 | si:dkey-14d8.6 | -0.069 |
| dusp2 | 0.109 | pdia2 | -0.068 |