BCL2 associated athanogene 5
ZFIN 
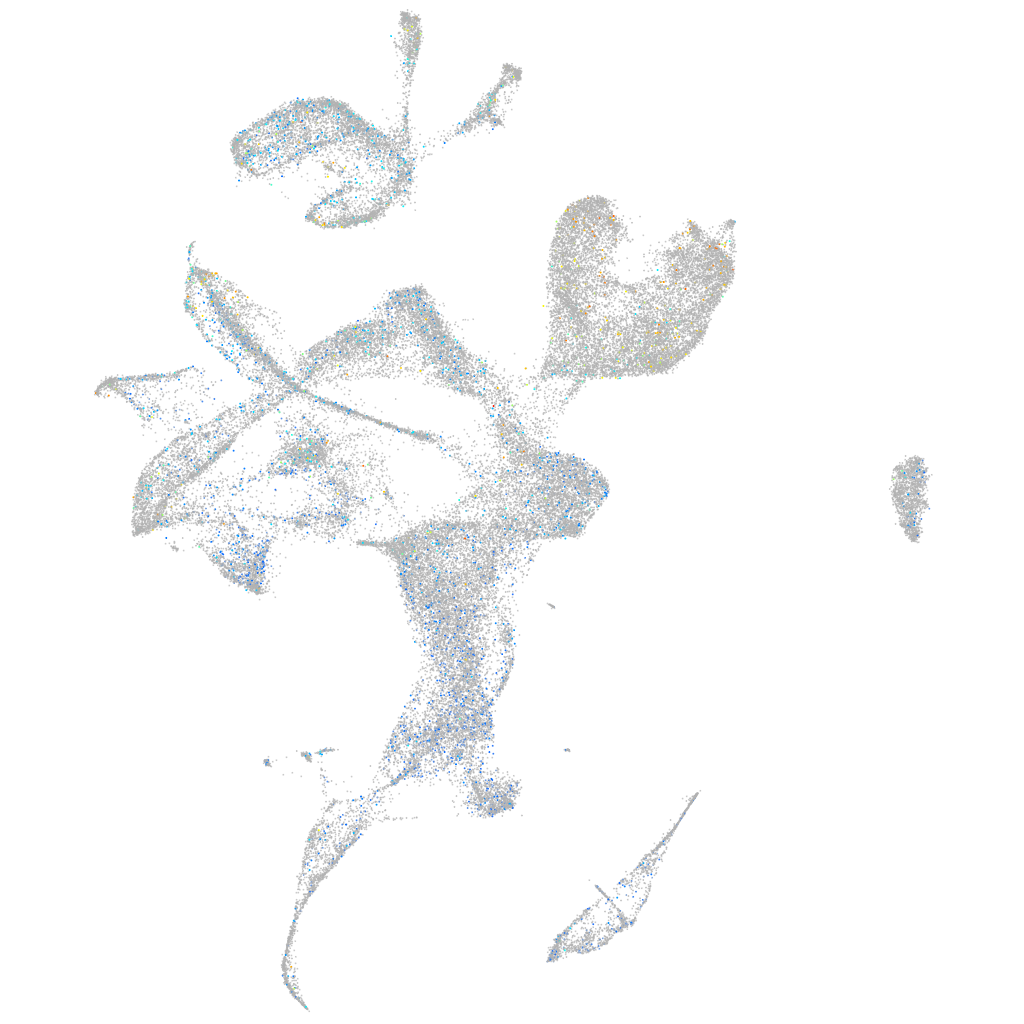
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cacna1aa | 0.050 | hmgb2a | -0.037 |
| rbfox1 | 0.050 | chaf1a | -0.022 |
| sncb | 0.049 | slbp | -0.021 |
| eno2 | 0.049 | stm | -0.021 |
| gabrb4 | 0.049 | pcna | -0.020 |
| ywhag2 | 0.048 | lig1 | -0.019 |
| rtn1b | 0.047 | shisa2a | -0.019 |
| syt2a | 0.047 | si:ch211-152c2.3 | -0.019 |
| stx1b | 0.047 | neurod4 | -0.018 |
| sv2a | 0.046 | krcp | -0.018 |
| SPATA48 | 0.046 | rrm1 | -0.018 |
| elavl3 | 0.045 | rps29 | -0.017 |
| cdk5r1b | 0.045 | mcm5 | -0.017 |
| snap25a | 0.045 | asf1ba | -0.017 |
| maptb | 0.045 | stmn1a | -0.017 |
| gnao1a | 0.045 | cdc6 | -0.017 |
| stmn2a | 0.045 | msna | -0.017 |
| zfhx3 | 0.044 | psmc3ip | -0.017 |
| stxbp1a | 0.044 | gadd45gb.1 | -0.017 |
| LOC100536949 | 0.044 | zgc:110540 | -0.016 |
| nrxn2a | 0.043 | dek | -0.016 |
| LOC108183650 | 0.043 | hes2.2 | -0.016 |
| grin1a | 0.043 | mcm6 | -0.016 |
| si:dkeyp-117h8.2 | 0.042 | rpa3 | -0.016 |
| myo10l1 | 0.042 | gins3 | -0.016 |
| gng3 | 0.042 | lbr | -0.016 |
| zgc:65894 | 0.041 | XLOC-042899 | -0.016 |
| elavl4 | 0.041 | tuba8l4 | -0.016 |
| ppp1r14ba | 0.041 | si:ch211-113a14.18 | -0.015 |
| zgc:153426 | 0.041 | hes6 | -0.015 |
| si:ch73-119p20.1 | 0.041 | dut | -0.015 |
| syt1a | 0.041 | tuba1b | -0.015 |
| si:ch211-129p13.1 | 0.040 | dhx32b | -0.015 |
| atp2b3b | 0.040 | CABZ01005379.1 | -0.015 |
| vsnl1b | 0.040 | dct | -0.015 |