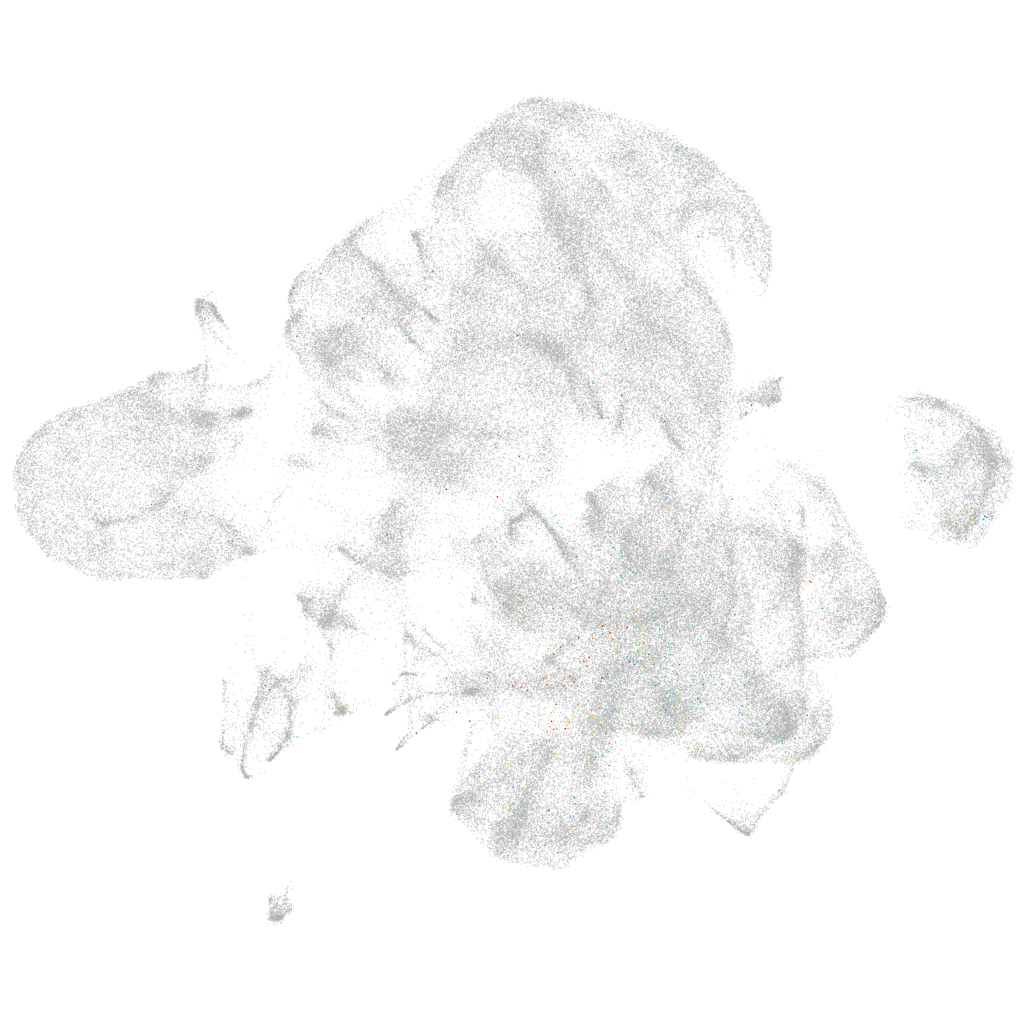
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 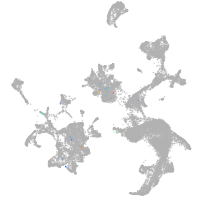 |
pronephros  |
neural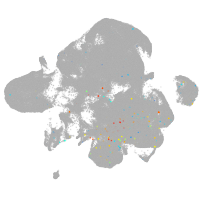 |
spinal cord / glia |
eye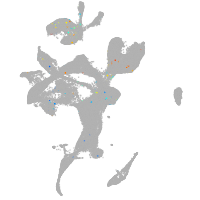 |
taste / olfactory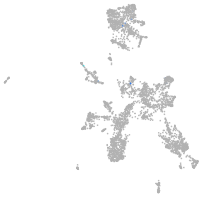 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103910432 | 0.070 | hmga1a | -0.050 |
| cd4-1 | 0.064 | eef1a1l1 | -0.049 |
| roraa | 0.063 | hmgb2a | -0.045 |
| jak3 | 0.058 | rps28 | -0.042 |
| ywhag2 | 0.056 | rplp2l | -0.042 |
| LOC103910375 | 0.055 | rps12 | -0.041 |
| ddit3 | 0.053 | rpsa | -0.039 |
| snap25a | 0.052 | rps6 | -0.038 |
| gng3 | 0.052 | rpl7a | -0.038 |
| atp6v1e1b | 0.050 | si:dkey-151g10.6 | -0.037 |
| sncgb | 0.050 | khdrbs1a | -0.037 |
| snap25b | 0.050 | hmgb2b | -0.037 |
| sncb | 0.049 | rpl29 | -0.037 |
| gpm6ab | 0.049 | rpl12 | -0.035 |
| atp6v0cb | 0.049 | rpl22l1 | -0.035 |
| eno2 | 0.048 | rpl11 | -0.035 |
| stmn2a | 0.048 | rps19 | -0.035 |
| gapdhs | 0.047 | id1 | -0.035 |
| stmn1b | 0.047 | rpl8 | -0.034 |
| tmsb2 | 0.047 | rpl23a | -0.034 |
| map1aa | 0.046 | tuba8l4 | -0.034 |
| ywhag1 | 0.046 | rpl4 | -0.033 |
| si:dkeyp-72g9.4 | 0.045 | rpl3 | -0.033 |
| ctsbb | 0.045 | pcna | -0.033 |
| atp6v1g1 | 0.045 | rps9 | -0.033 |
| rnasekb | 0.045 | ppiaa | -0.033 |
| calm1a | 0.045 | rps5 | -0.033 |
| vamp2 | 0.045 | rps20 | -0.033 |
| fam49al | 0.045 | tpt1 | -0.033 |
| ppdpfb | 0.044 | rpl35a | -0.032 |
| necap1 | 0.044 | cx43.4 | -0.032 |
| atf5b | 0.044 | rpl15 | -0.032 |
| pacsin1a | 0.044 | rpl18a | -0.032 |
| stx1b | 0.044 | cirbpa | -0.032 |
| gpm6aa | 0.044 | rpl36a | -0.032 |