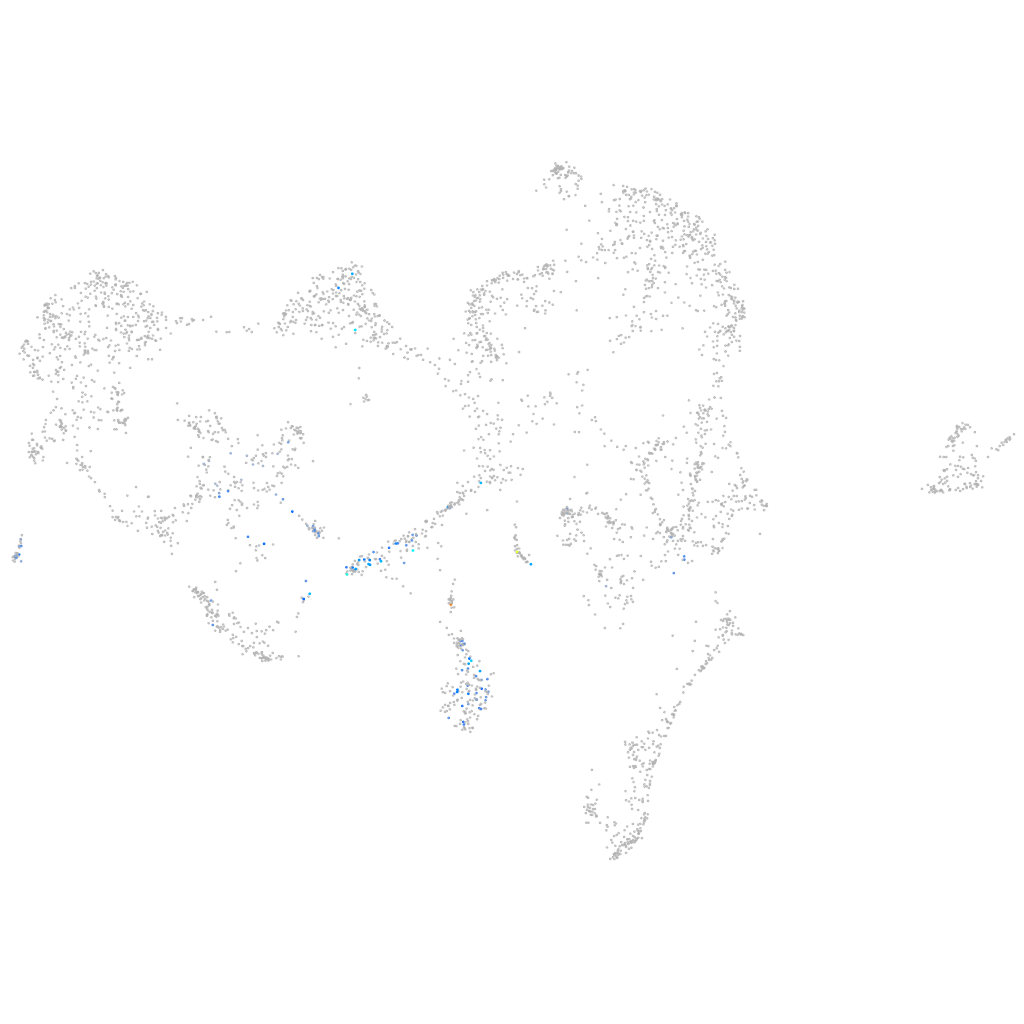
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 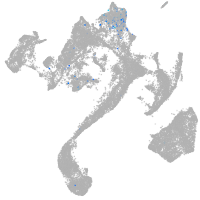 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory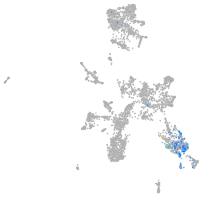 |
otic / lateral line |
epidermis |
periderm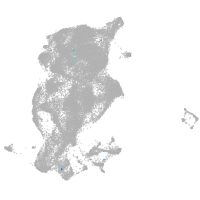 |
fin |
ionocytes / mucous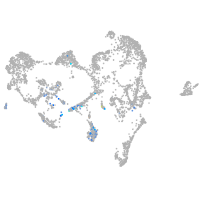 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ceacam1 | 0.337 | rplp1 | -0.128 |
| ca15a | 0.323 | krt91 | -0.112 |
| zgc:193726 | 0.312 | rps21 | -0.108 |
| atp6v0a1a | 0.311 | actb1 | -0.101 |
| cox5b2 | 0.310 | rps19 | -0.101 |
| XLOC-014886 | 0.309 | eef1a1l1 | -0.100 |
| slc4a1b | 0.308 | rps13 | -0.096 |
| cacng5a | 0.302 | BX000438.2 | -0.095 |
| XLOC-043659 | 0.299 | aldob | -0.092 |
| clec11a | 0.290 | lgals3b | -0.092 |
| si:ch211-147d7.5 | 0.290 | junba | -0.088 |
| atp6v1aa | 0.290 | agr2 | -0.086 |
| cnnm2a | 0.284 | rps24 | -0.086 |
| atp6v0ca | 0.281 | hmgn2 | -0.086 |
| atp6v1g1 | 0.280 | rps15a | -0.086 |
| si:dkey-192d15.2 | 0.280 | fxyd6l | -0.085 |
| rnaseka | 0.274 | her6 | -0.084 |
| atp6ap2 | 0.272 | si:dkey-151g10.6 | -0.083 |
| atp6v1ba | 0.269 | rps10 | -0.083 |
| atp6v1e1b | 0.267 | spaca4l | -0.083 |
| atp6v0d1 | 0.262 | rps26l | -0.081 |
| atp6ap1b | 0.262 | CR383676.1 | -0.080 |
| atp6v0b | 0.261 | ncl | -0.079 |
| si:ch211-214j24.14 | 0.261 | rpl28 | -0.079 |
| ca2 | 0.260 | dap1b | -0.079 |
| sat1a.1 | 0.259 | rps27.1 | -0.079 |
| slc31a2 | 0.255 | cd63 | -0.078 |
| serpinc1 | 0.252 | eif4ebp3l | -0.078 |
| foxi3a | 0.252 | rpl36 | -0.078 |
| rhcgb | 0.246 | muc5.1 | -0.077 |
| atp6v1h | 0.246 | rpl23 | -0.077 |
| trpm7 | 0.243 | rpl14 | -0.075 |
| clcn2b | 0.242 | krt5 | -0.075 |
| atp6v1f | 0.242 | rpl7a | -0.075 |
| si:ch211-195o20.7 | 0.241 | eef2b | -0.074 |