Other cell groups


all cells |
blastomeres |
PGCs |
endoderm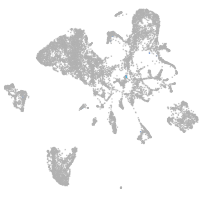 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural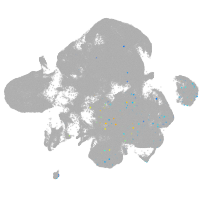 |
spinal cord / glia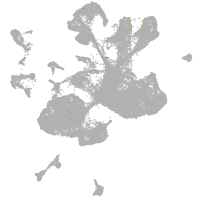 |
eye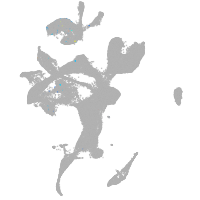 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR394546.3 | 0.122 | hmgb2a | -0.056 |
| pax10 | 0.114 | msi1 | -0.034 |
| si:dkey-269p2.1 | 0.109 | msna | -0.033 |
| LOC103909566 | 0.105 | tuba8l4 | -0.032 |
| tfap2a | 0.104 | ahcy | -0.031 |
| slc32a1 | 0.103 | nutf2l | -0.030 |
| mir181b-3 | 0.099 | tuba8l | -0.030 |
| tfap2e | 0.098 | pcna | -0.029 |
| cabp1b | 0.091 | ccnd1 | -0.029 |
| tfap2b | 0.091 | chaf1a | -0.029 |
| celf5a | 0.091 | stmn1a | -0.029 |
| tfap2c | 0.089 | mki67 | -0.029 |
| mtnr1c | 0.089 | mdka | -0.029 |
| stxbp1a | 0.085 | her15.1 | -0.029 |
| LOC101886114 | 0.085 | eef1da | -0.028 |
| AL627126.1 | 0.084 | crx | -0.028 |
| elavl3 | 0.082 | ccna2 | -0.028 |
| carmil2 | 0.073 | rrm2 | -0.027 |
| syt1a | 0.072 | rrm1 | -0.027 |
| meis2a | 0.072 | lbr | -0.027 |
| si:ch73-290k24.5 | 0.072 | mcm7 | -0.027 |
| lta | 0.070 | selenoh | -0.027 |
| plxna2 | 0.069 | zgc:110425 | -0.027 |
| mir181b-1 | 0.069 | lig1 | -0.027 |
| pax6a | 0.068 | histh1l | -0.026 |
| cplx3b | 0.067 | rpa3 | -0.026 |
| rpp25a | 0.067 | mibp | -0.026 |
| meis2b | 0.066 | tubb4b | -0.026 |
| basp1 | 0.066 | syt5b | -0.026 |
| XLOC-031260 | 0.066 | dut | -0.025 |
| cacng4a | 0.066 | otx5 | -0.025 |
| XLOC-004681 | 0.066 | aurkb | -0.025 |
| stx1b | 0.066 | id1 | -0.025 |
| gad2 | 0.065 | mad2l1 | -0.025 |
| CR450824.2 | 0.065 | dek | -0.025 |




