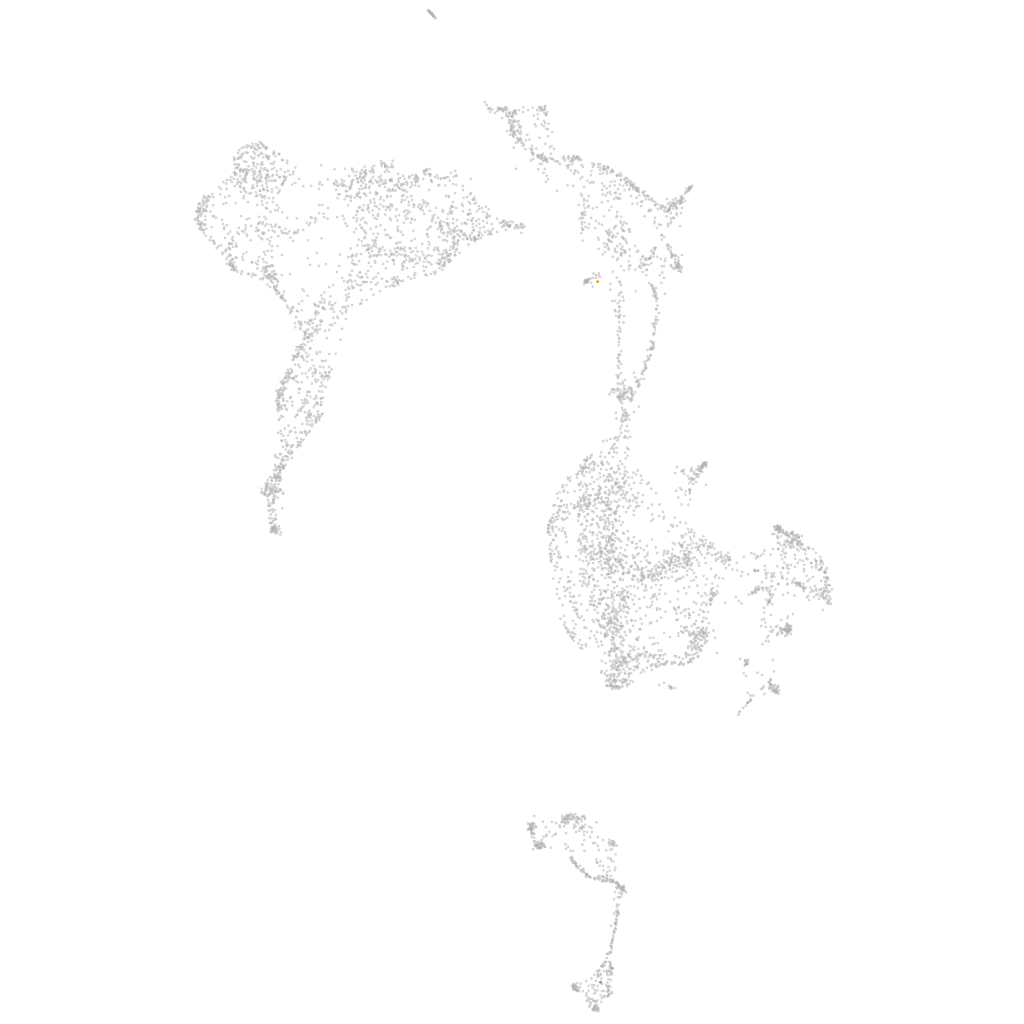
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr132b | 0.818 | ybx1 | -0.054 |
| rap1gap2b | 0.670 | pabpc1a | -0.049 |
| onecut3a | 0.666 | rps7 | -0.047 |
| rragd | 0.486 | rpl34 | -0.042 |
| cers4b | 0.486 | rps6 | -0.039 |
| plac8l1 | 0.476 | eef2b | -0.038 |
| npsn | 0.466 | rps27.2 | -0.035 |
| plch1 | 0.457 | rpl9 | -0.031 |
| XLOC-041945 | 0.412 | qkia | -0.030 |
| clec19a | 0.404 | ran | -0.029 |
| exoc3l2a | 0.362 | hmgn7 | -0.029 |
| ponzr4 | 0.349 | ppib | -0.028 |
| hdac7a | 0.328 | hnrnpaba | -0.028 |
| si:ch73-52f15.5 | 0.325 | serf2 | -0.028 |
| cryba1l1 | 0.313 | rpl27a | -0.027 |
| gfpt2 | 0.312 | ndufa4l | -0.027 |
| crybb1l2 | 0.308 | xbp1 | -0.027 |
| LOC556707 | 0.266 | cirbpa | -0.026 |
| brip1 | 0.251 | cox6b2 | -0.026 |
| XLOC-016167 | 0.244 | eif3f | -0.026 |
| msh3 | 0.241 | btf3 | -0.026 |
| lmx1bb | 0.238 | calm2b | -0.025 |
| C12orf75 | 0.237 | hnrnpa0b | -0.025 |
| tnni2b.1 | 0.228 | msna | -0.025 |
| h6pd | 0.221 | hmga1a | -0.024 |
| syngr3b | 0.206 | hmgn6 | -0.024 |
| CT956002.2 | 0.204 | fkbp1aa | -0.024 |
| auts2b | 0.196 | si:ch73-281n10.2 | -0.024 |
| sqor | 0.191 | syncrip | -0.024 |
| clp1 | 0.185 | rbm8a | -0.024 |
| slc7a6 | 0.185 | ddx5 | -0.024 |
| frrs1b | 0.180 | si:ch211-288g17.3 | -0.024 |
| hsp90aa1.1 | 0.177 | cx43.4 | -0.024 |
| shprh | 0.175 | slc25a3b | -0.023 |
| XLOC-013964 | 0.175 | hp1bp3 | -0.023 |