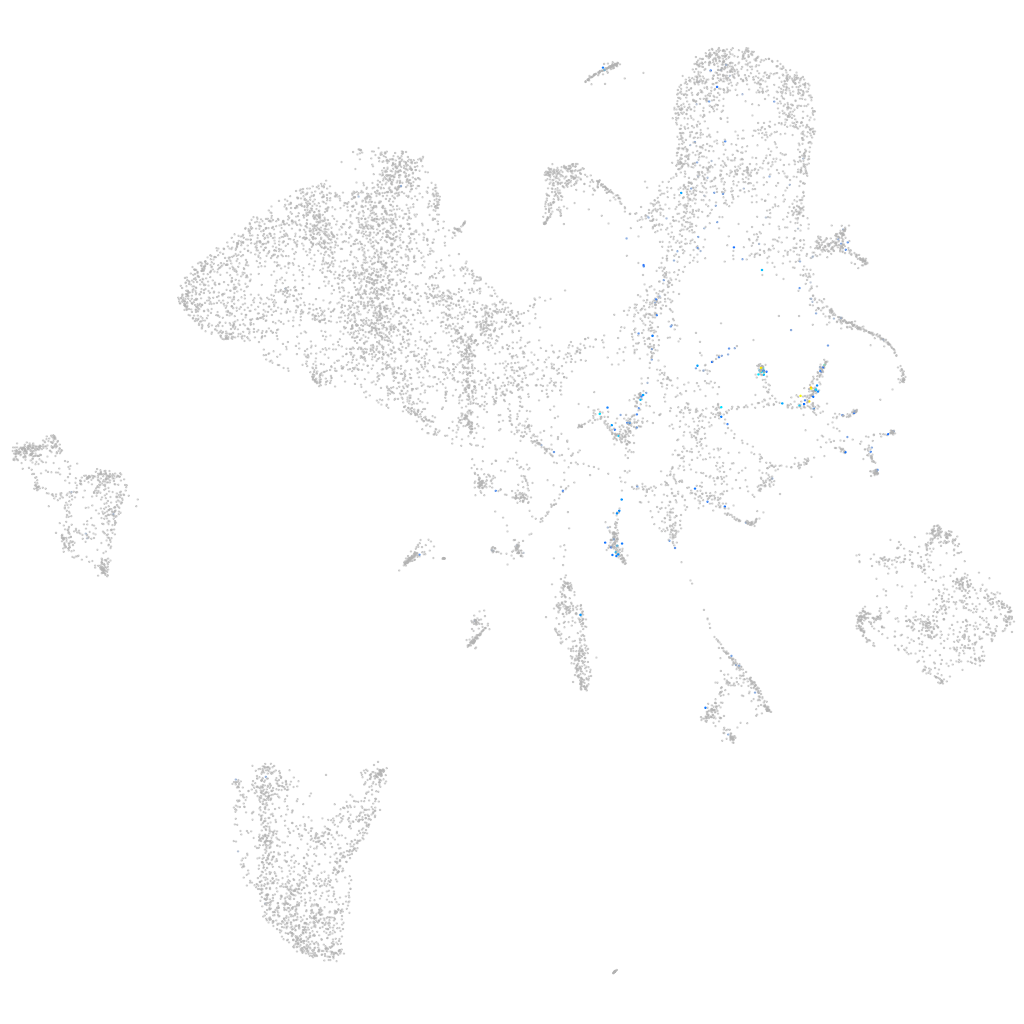
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccka | 0.279 | bhmt | -0.090 |
| si:zfos-2372e4.1 | 0.273 | gatm | -0.086 |
| cckb | 0.241 | gamt | -0.084 |
| pax4 | 0.215 | mat1a | -0.076 |
| scg3 | 0.211 | apoc1 | -0.076 |
| TCIM (1 of many) | 0.208 | aqp12 | -0.075 |
| capsla | 0.202 | apoa1b | -0.075 |
| nfatc2b | 0.196 | agxtb | -0.074 |
| insm1b | 0.195 | fabp3 | -0.074 |
| rasd4 | 0.190 | hdlbpa | -0.073 |
| XLOC-013106 | 0.189 | sec61a1 | -0.073 |
| rprma | 0.186 | tfa | -0.072 |
| mir7a-1 | 0.185 | gapdh | -0.071 |
| CR457444.9 | 0.182 | ahcy | -0.071 |
| taar13b | 0.182 | grhprb | -0.071 |
| CT990561.2 | 0.180 | apoa4b.1 | -0.071 |
| vat1 | 0.175 | apoa2 | -0.070 |
| fsip1 | 0.174 | mgst1.2 | -0.070 |
| si:busm1-57f23.1 | 0.172 | afp4 | -0.069 |
| kcnt2 | 0.170 | fbp1b | -0.068 |
| gfra1a | 0.169 | serpina1 | -0.068 |
| egr4 | 0.168 | ttc36 | -0.068 |
| anxa4 | 0.164 | cx32.3 | -0.067 |
| npy2rl | 0.164 | agxta | -0.067 |
| elovl1a | 0.164 | kng1 | -0.067 |
| rem2 | 0.163 | serpina1l | -0.067 |
| dll4 | 0.162 | ambp | -0.066 |
| fev | 0.162 | fabp10a | -0.066 |
| s100a1 | 0.160 | rbp2b | -0.066 |
| FADS6 | 0.159 | si:dkey-16p21.8 | -0.066 |
| gfra3 | 0.159 | apom | -0.066 |
| neurod1 | 0.158 | uox | -0.065 |
| ush1c | 0.155 | ces2 | -0.065 |
| slc8a1b | 0.153 | hpda | -0.065 |
| pyyb | 0.153 | fgb | -0.065 |