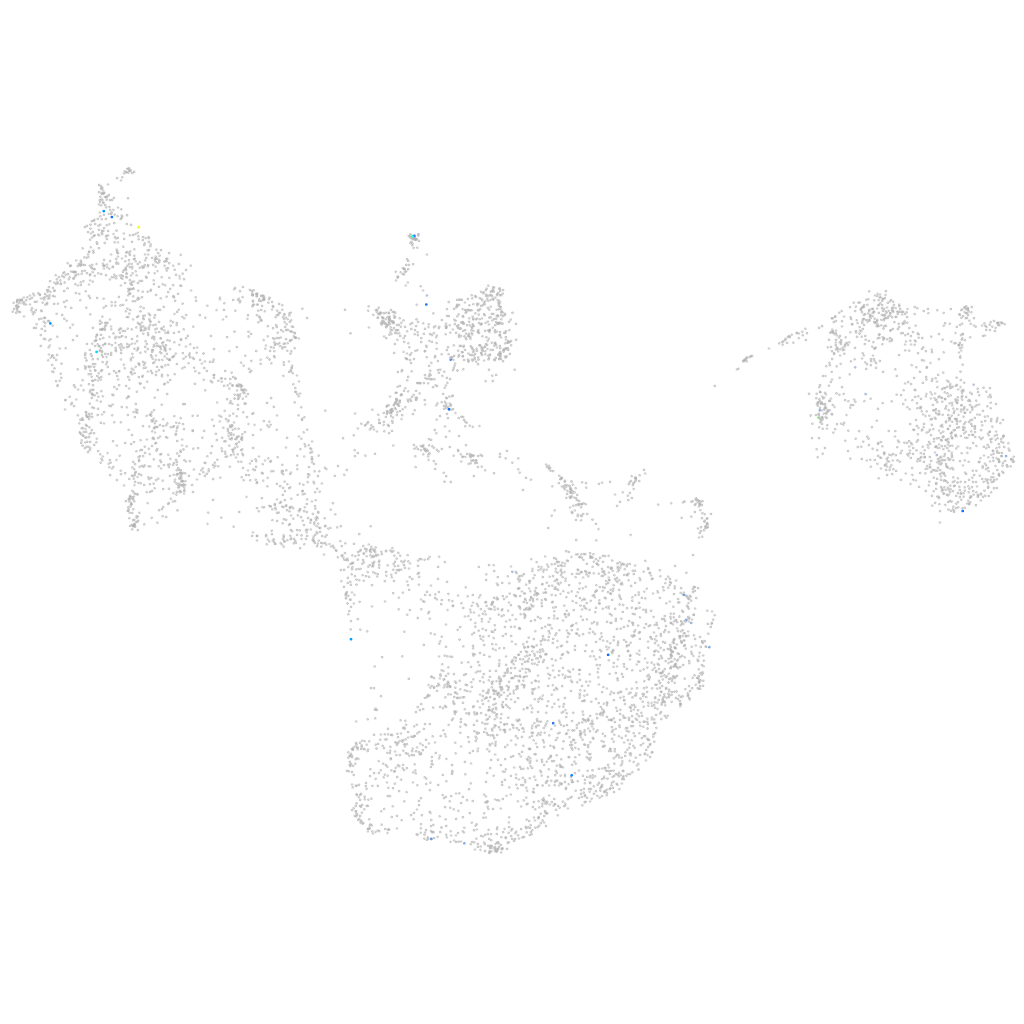
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm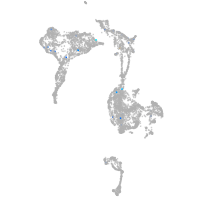 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 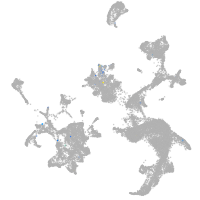 |
pronephros  |
neural |
spinal cord / glia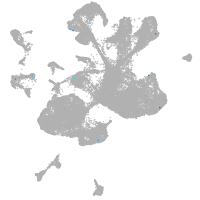 |
eye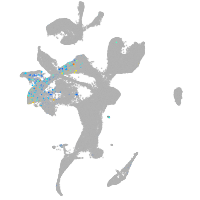 |
taste / olfactory |
otic / lateral line |
epidermis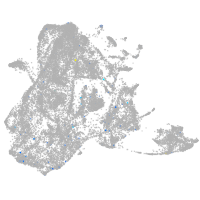 |
periderm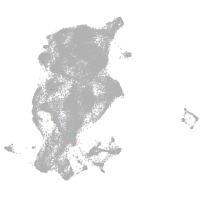 |
fin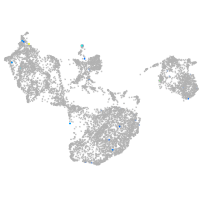 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC100331665 | 0.375 | cox5ab | -0.026 |
| lgals9l4 | 0.375 | mrpl54 | -0.024 |
| fosl1b | 0.366 | brms1la | -0.024 |
| si:dkey-14o18.2 | 0.358 | lsm3 | -0.024 |
| cldn2 | 0.334 | oip5-as1 | -0.023 |
| nrtn | 0.309 | emd | -0.023 |
| zgc:193593 | 0.280 | mgst3a | -0.023 |
| ptger4c | 0.267 | nifk | -0.023 |
| ngs | 0.252 | mt-nd3 | -0.023 |
| ttc8.1 | 0.245 | ppp1caa | -0.022 |
| ptafr | 0.242 | cxcl14 | -0.022 |
| uts1 | 0.235 | fam53b | -0.022 |
| si:dkey-26i13.8 | 0.221 | glrx | -0.022 |
| clul1 | 0.214 | srsf7a | -0.022 |
| camk1ga | 0.206 | fkbp1aa | -0.021 |
| LOC101885444 | 0.206 | fkbp3 | -0.021 |
| CR936977.1 | 0.201 | ndufv3 | -0.021 |
| glis3 | 0.199 | prr13 | -0.021 |
| rgs9a | 0.194 | rbbp6 | -0.020 |
| pde5aa | 0.190 | zmp:0000000846 | -0.020 |
| inaa | 0.178 | tram2 | -0.020 |
| cnga3b | 0.176 | caspb | -0.020 |
| eno4 | 0.174 | DHDH | -0.020 |
| rp1 | 0.172 | calm2a | -0.020 |
| CR847898.4 | 0.168 | arpc3 | -0.020 |
| pde6h | 0.163 | st13 | -0.020 |
| guca1c | 0.162 | si:dkey-42i9.6 | -0.020 |
| lrit2 | 0.161 | prelid1a | -0.020 |
| si:cabz01023700.1 | 0.161 | ptmaa | -0.020 |
| impg1a | 0.160 | eif4a1b | -0.019 |
| XLOC-043546 | 0.160 | yipf6 | -0.019 |
| LOC108179355 | 0.157 | CU694197.1 | -0.019 |
| LOC110438150 | 0.157 | si:dkey-52l18.4 | -0.019 |
| TC2N | 0.155 | wnt5b | -0.019 |
| BX323861.3 | 0.153 | lasp1 | -0.019 |