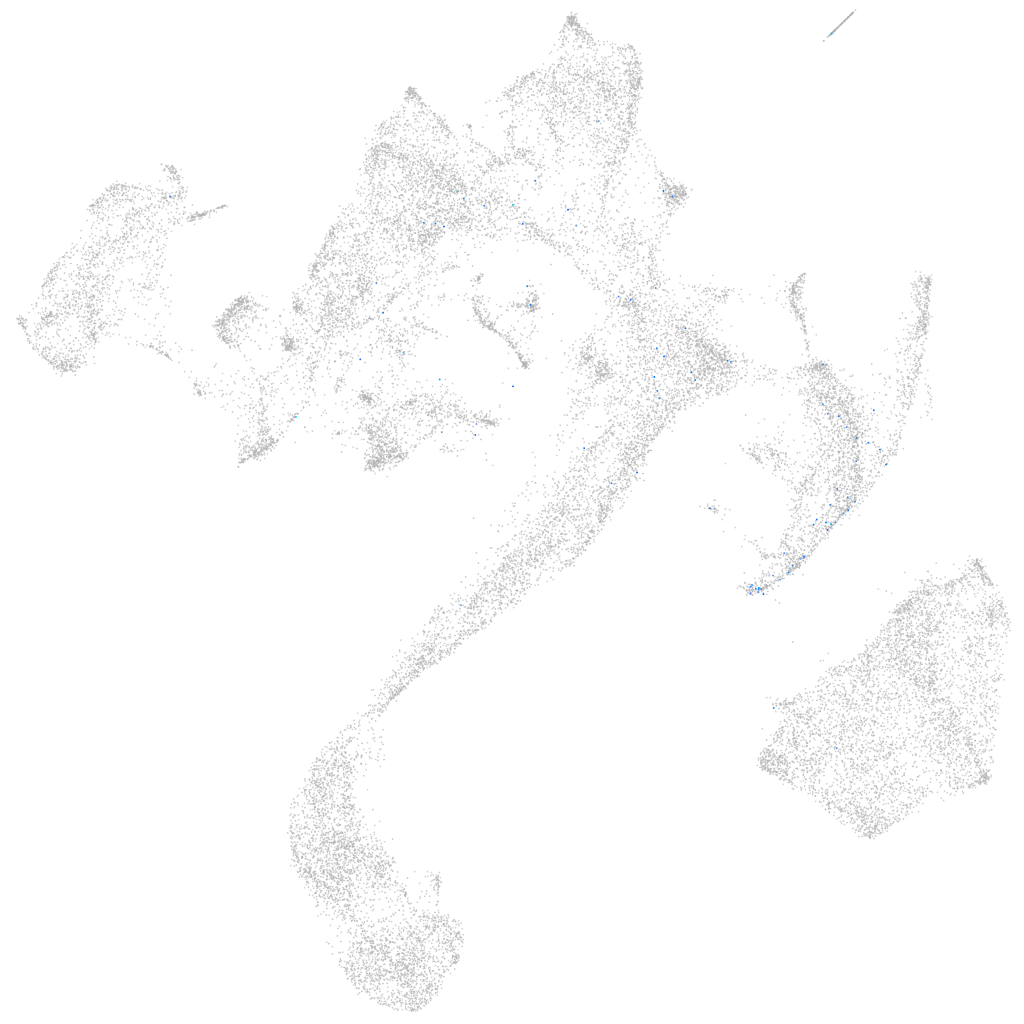
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 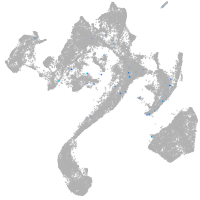 |
mural cells / non-skeletal muscle 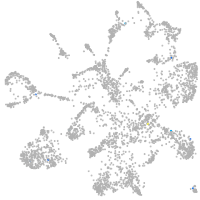 |
mesenchyme 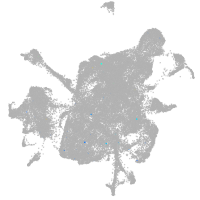 |
hematopoietic / vasculature 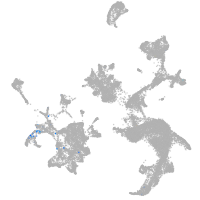 |
pronephros 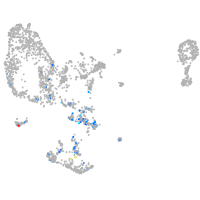 |
neural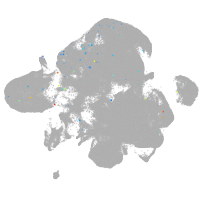 |
spinal cord / glia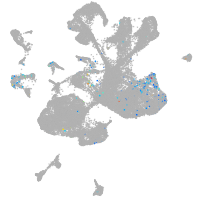 |
eye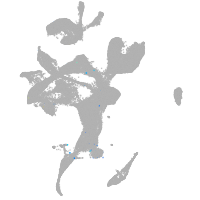 |
taste / olfactory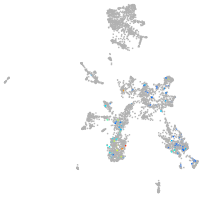 |
otic / lateral line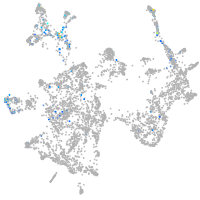 |
epidermis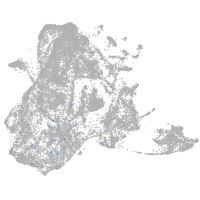 |
periderm |
fin |
ionocytes / mucous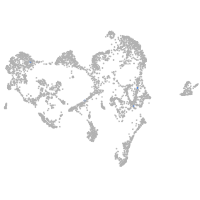 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX323994.1 | 0.192 | actc1b | -0.028 |
| CR927456.1 | 0.140 | tpi1b | -0.027 |
| BX569790.1 | 0.106 | aldoab | -0.025 |
| pimr92 | 0.104 | ckma | -0.025 |
| LOC101882982 | 0.104 | tmem38a | -0.024 |
| tbpl2 | 0.094 | atp2a1 | -0.024 |
| ak7b | 0.093 | ckmb | -0.024 |
| kcnh4b | 0.090 | tmem182a | -0.024 |
| si:dkey-71b5.6 | 0.090 | ak1 | -0.024 |
| CR854988.1 | 0.088 | acta1b | -0.024 |
| si:ch211-220f16.1 | 0.087 | si:dkey-16p21.8 | -0.024 |
| dhx58 | 0.087 | eno1a | -0.023 |
| dnaaf3 | 0.087 | gamt | -0.023 |
| cxcr2 | 0.082 | txlnba | -0.023 |
| CU929359.1 | 0.082 | pgam2 | -0.023 |
| si:ch211-5k11.8 | 0.081 | tpma | -0.023 |
| XLOC-032267 | 0.081 | acta1a | -0.022 |
| CU207217.1 | 0.079 | gapdh | -0.022 |
| shisa6 | 0.078 | tnnc2 | -0.022 |
| cfap126 | 0.075 | srl | -0.022 |
| cfap46 | 0.074 | mylpfa | -0.022 |
| si:ch73-343l4.8 | 0.073 | actn3a | -0.022 |
| CR376741.6 | 0.073 | rtn3 | -0.021 |
| kif17 | 0.073 | neb | -0.021 |
| daw1 | 0.072 | pvalb2 | -0.021 |
| cdc25d | 0.072 | pkmb | -0.021 |
| wdr66 | 0.071 | tmod4 | -0.021 |
| LOC103908877 | 0.068 | ttn.2 | -0.021 |
| dnah9 | 0.068 | si:dkey-211f22.5 | -0.021 |
| her12 | 0.068 | cox17 | -0.021 |
| sall4 | 0.068 | dhrs7cb | -0.021 |
| LOC108181470 | 0.067 | ank1a | -0.021 |
| lrp2bp | 0.067 | atp1a2a | -0.021 |
| BX901907.4 | 0.066 | CABZ01078594.1 | -0.021 |
| LOC108179692 | 0.066 | ldb3a | -0.020 |