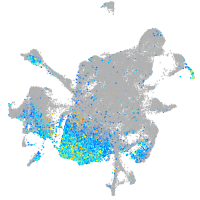
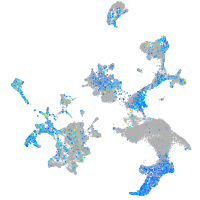
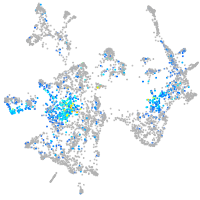
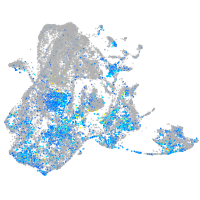
aurora kinase B
ZFIN 
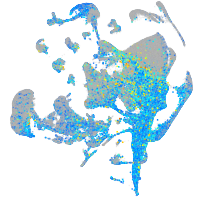
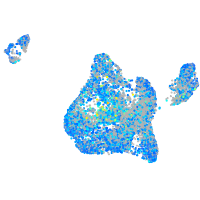
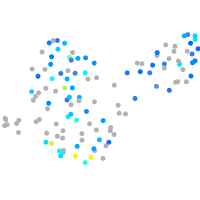
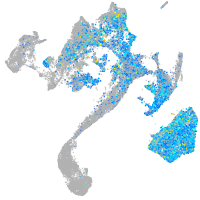
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdk1 | 0.647 | actc1b | -0.394 |
| mad2l1 | 0.641 | ttn.2 | -0.358 |
| mki67 | 0.634 | ttn.1 | -0.351 |
| tpx2 | 0.631 | aldoab | -0.347 |
| ube2c | 0.623 | ak1 | -0.344 |
| plk1 | 0.609 | tmem38a | -0.339 |
| ccnb1 | 0.600 | atp2a1 | -0.339 |
| top2a | 0.593 | ckmb | -0.336 |
| si:ch211-69g19.2 | 0.590 | ckma | -0.335 |
| nusap1 | 0.586 | tnnc2 | -0.330 |
| kpna2 | 0.580 | acta1b | -0.322 |
| ccna2 | 0.572 | desma | -0.322 |
| cdca8 | 0.569 | pabpc4 | -0.320 |
| cdc20 | 0.558 | acta1a | -0.319 |
| hmgb2a | 0.545 | mybphb | -0.318 |
| tubb2b | 0.538 | tpma | -0.316 |
| kif11 | 0.534 | neb | -0.315 |
| aspm | 0.533 | atp5if1b | -0.315 |
| cenpf | 0.533 | mylpfa | -0.314 |
| kifc1 | 0.533 | srl | -0.314 |
| aurka | 0.532 | ldb3a | -0.314 |
| kif23 | 0.527 | gamt | -0.309 |
| birc5a | 0.525 | eno1a | -0.308 |
| cks1b | 0.520 | CABZ01078594.1 | -0.306 |
| dlgap5 | 0.517 | cav3 | -0.304 |
| kmt5ab | 0.502 | vdac3 | -0.303 |
| tuba8l4 | 0.501 | ldb3b | -0.299 |
| tacc3 | 0.489 | gapdh | -0.298 |
| prc1a | 0.487 | klhl41b | -0.296 |
| armc1l | 0.486 | actn3b | -0.295 |
| anp32b | 0.486 | nme2b.2 | -0.292 |
| hmgb2b | 0.482 | myl1 | -0.290 |
| lbr | 0.482 | eno3 | -0.288 |
| anp32a | 0.480 | si:ch73-367p23.2 | -0.288 |
| ccnb3 | 0.478 | txlnbb | -0.287 |