ATPase H+ transporting V0 subunit e2
ZFIN 
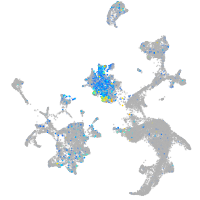
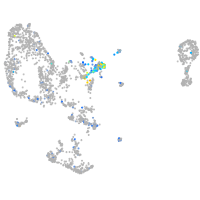
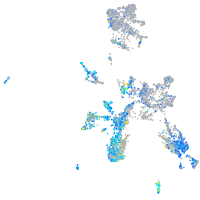
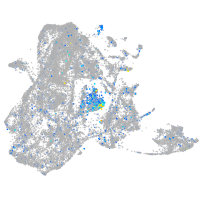
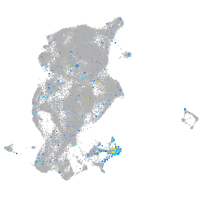
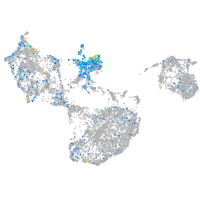
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.409 | ahcy | -0.181 |
| neurod1 | 0.381 | gapdh | -0.171 |
| syt1a | 0.372 | gamt | -0.164 |
| pcsk1nl | 0.357 | aldob | -0.160 |
| aldocb | 0.350 | gatm | -0.154 |
| atp6v0cb | 0.343 | mat1a | -0.142 |
| mir7a-1 | 0.341 | aldh6a1 | -0.139 |
| rnasekb | 0.340 | fbp1b | -0.138 |
| snap25a | 0.338 | cx32.3 | -0.133 |
| vamp2 | 0.332 | agxtb | -0.132 |
| scgn | 0.330 | gstt1a | -0.132 |
| vat1 | 0.323 | aqp12 | -0.131 |
| gapdhs | 0.322 | bhmt | -0.128 |
| gnao1a | 0.321 | apoa4b.1 | -0.128 |
| pax6b | 0.314 | gstr | -0.127 |
| insm1a | 0.314 | eno3 | -0.127 |
| tspan7b | 0.313 | scp2a | -0.126 |
| id4 | 0.310 | aldh7a1 | -0.124 |
| insm1b | 0.310 | agxta | -0.123 |
| gpc1a | 0.306 | hspe1 | -0.122 |
| kcnj11 | 0.304 | nupr1b | -0.122 |
| gdi1 | 0.298 | g6pca.2 | -0.121 |
| cpe | 0.298 | gnmt | -0.121 |
| si:dkey-153k10.9 | 0.296 | shmt1 | -0.121 |
| slc35g2a | 0.293 | mgst1.2 | -0.120 |
| eno1a | 0.293 | hdlbpa | -0.120 |
| zgc:101731 | 0.293 | apoa1b | -0.120 |
| c2cd4a | 0.291 | pnp4b | -0.120 |
| egr4 | 0.290 | glud1b | -0.119 |
| dpysl2b | 0.290 | apoc2 | -0.119 |
| rims2a | 0.288 | ttc36 | -0.119 |
| cnrip1a | 0.287 | si:dkey-16p21.8 | -0.118 |
| camk2n1a | 0.285 | nipsnap3a | -0.117 |
| nkx2.2a | 0.284 | rps20 | -0.117 |
| fev | 0.284 | slc16a1b | -0.116 |