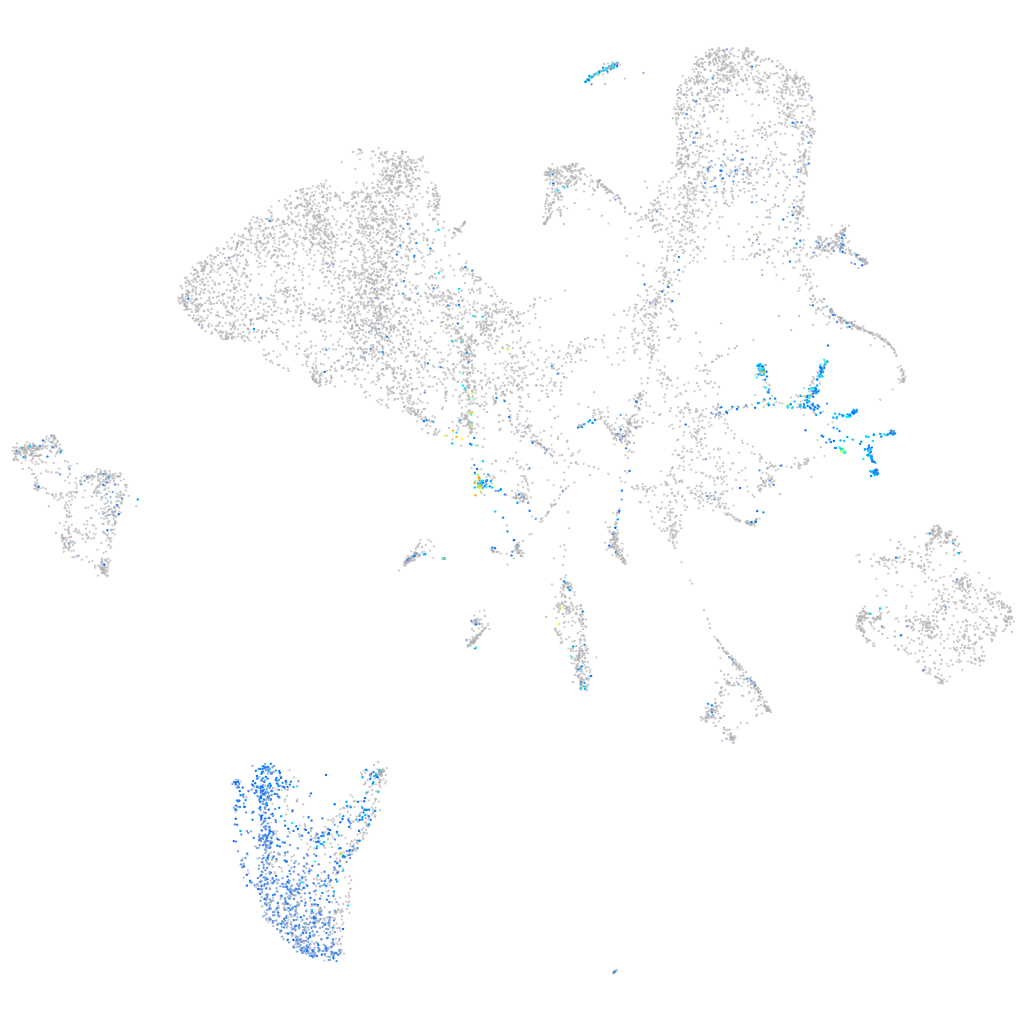
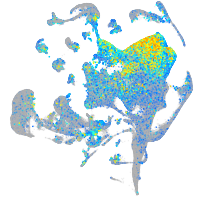
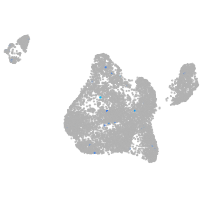
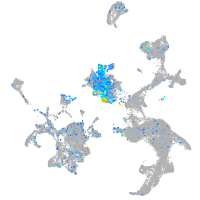
ATPase H+ transporting V0 subunit cb
ZFIN 
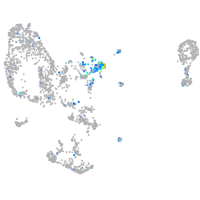
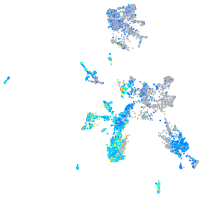
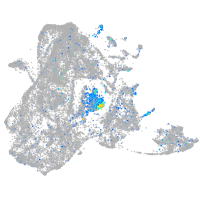
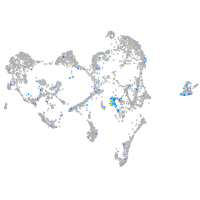
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1a | 0.472 | gstr | -0.263 |
| scg3 | 0.470 | aldob | -0.255 |
| syt1a | 0.423 | gstt1a | -0.254 |
| neurod1 | 0.423 | apoa4b.1 | -0.251 |
| scgn | 0.420 | prdx2 | -0.242 |
| pcsk1nl | 0.416 | gstp1 | -0.241 |
| ptprna | 0.397 | apoc2 | -0.240 |
| pax6b | 0.382 | ugt1a7 | -0.239 |
| rnasekb | 0.380 | mgst1.2 | -0.236 |
| mir7a-1 | 0.379 | eno3 | -0.230 |
| slc1a4 | 0.374 | gpx1a | -0.229 |
| id4 | 0.369 | sdr16c5b | -0.229 |
| vamp2 | 0.366 | scp2a | -0.228 |
| kcnj11 | 0.359 | afp4 | -0.223 |
| aldocb | 0.356 | rdh1 | -0.218 |
| si:dkey-153k10.9 | 0.352 | apoc1 | -0.217 |
| pcsk1 | 0.348 | sult2st2 | -0.217 |
| tspan7b | 0.344 | apoa1b | -0.217 |
| atpv0e2 | 0.343 | si:dkey-16p21.8 | -0.216 |
| egr4 | 0.339 | dhrs9 | -0.216 |
| scg2a | 0.339 | tdo2a | -0.213 |
| rprmb | 0.337 | gsta.1 | -0.213 |
| gnao1a | 0.334 | aldh1l1 | -0.212 |
| snap25a | 0.333 | haao | -0.212 |
| insm1b | 0.330 | ftcd | -0.209 |
| insm1a | 0.330 | rmdn1 | -0.208 |
| pdia2 | 0.329 | apobb.1 | -0.207 |
| gpc1a | 0.327 | phyhd1 | -0.207 |
| gdi1 | 0.324 | gcshb | -0.206 |
| cnrip1a | 0.319 | etnppl | -0.202 |
| rims2a | 0.318 | msrb2 | -0.202 |
| cplx2l | 0.317 | aldh9a1a.1 | -0.202 |
| erp27 | 0.317 | cyp4v8 | -0.202 |
| vat1 | 0.316 | acmsd | -0.202 |
| cpe | 0.314 | igfbp2a | -0.201 |