ATPase phospholipid transporting 11A
ZFIN 
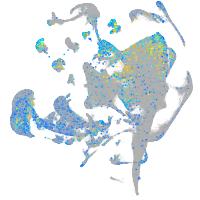
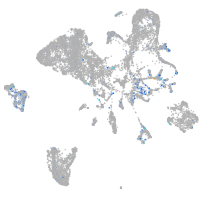
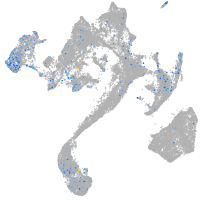
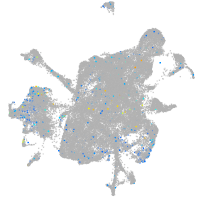
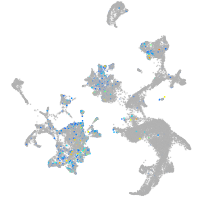
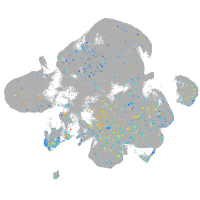
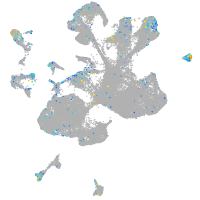
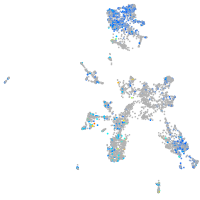
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110239 | 0.304 | CABZ01021592.1 | -0.185 |
| slc45a2 | 0.303 | ptmaa | -0.174 |
| SPAG9 | 0.302 | si:ch211-222l21.1 | -0.165 |
| agtrap | 0.291 | si:dkey-251i10.2 | -0.157 |
| tspan36 | 0.290 | tuba1c | -0.156 |
| rab32a | 0.283 | marcksl1a | -0.156 |
| atp6v0ca | 0.282 | gpm6aa | -0.141 |
| sptlc2a | 0.281 | si:ch73-1a9.3 | -0.137 |
| ctsba | 0.278 | elavl3 | -0.135 |
| BX571715.1 | 0.278 | hmgb1b | -0.133 |
| tyr | 0.274 | tmsb | -0.131 |
| canx | 0.267 | nova2 | -0.131 |
| atp6ap1b | 0.264 | stmn1b | -0.131 |
| psap | 0.263 | zc4h2 | -0.126 |
| pah | 0.261 | si:ch73-281n10.2 | -0.123 |
| trpm1b | 0.261 | hbbe1.3 | -0.123 |
| kita | 0.259 | pvalb2 | -0.120 |
| bace2 | 0.258 | fabp7a | -0.119 |
| lyst | 0.258 | hbae3 | -0.118 |
| col4a1 | 0.257 | cadm3 | -0.114 |
| CABZ01048402.2 | 0.256 | stmn1a | -0.113 |
| slc37a2 | 0.255 | actc1b | -0.111 |
| LOC103910009 | 0.254 | gng3 | -0.110 |
| myo5aa | 0.252 | hmgn2 | -0.109 |
| pcdh10a | 0.252 | hmgb3a | -0.108 |
| oca2 | 0.251 | CU467822.1 | -0.107 |
| lamp1a | 0.248 | epb41a | -0.107 |
| syngr1a | 0.246 | aldob | -0.106 |
| mc1r | 0.245 | tmeff1b | -0.106 |
| slc29a1a | 0.244 | pvalb1 | -0.106 |
| pnp4a | 0.241 | hbbe1.1 | -0.105 |
| rab38 | 0.241 | uraha | -0.104 |
| gpr143 | 0.241 | vamp2 | -0.103 |
| tlcd2 | 0.240 | dio3a | -0.103 |
| sypl2b | 0.237 | fez1 | -0.102 |