alpha tubulin acetyltransferase 1
ZFIN 
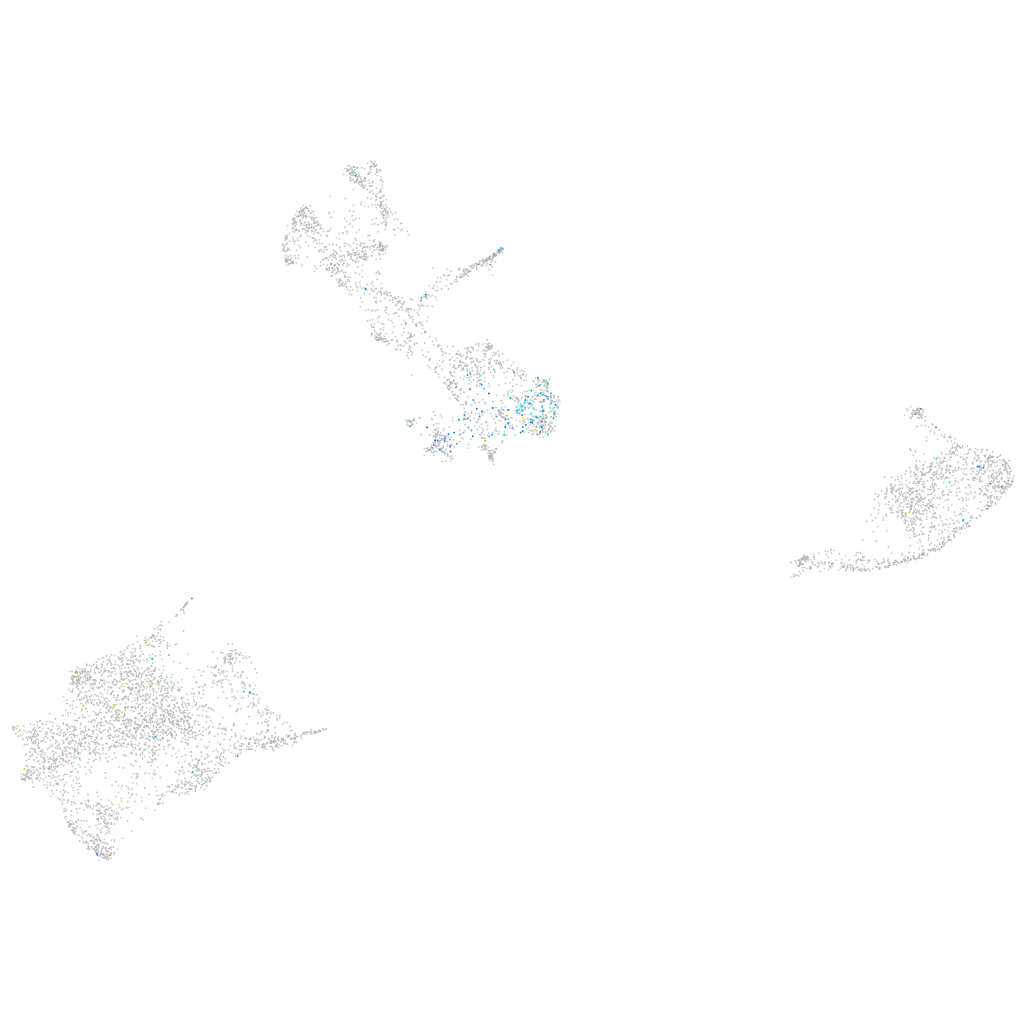
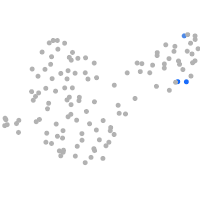
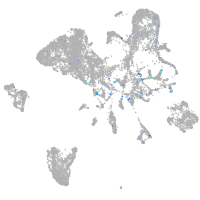
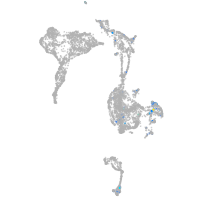
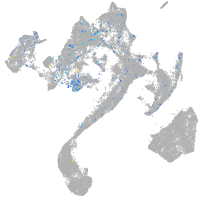
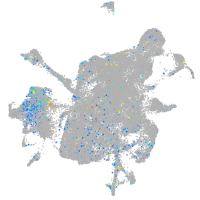
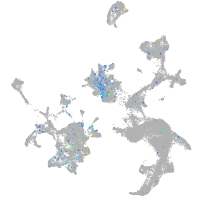
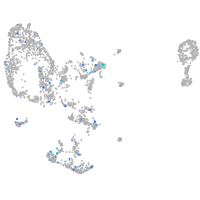
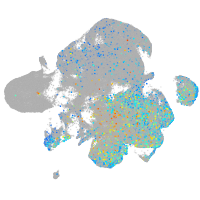
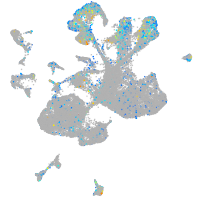
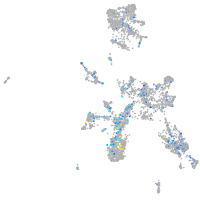
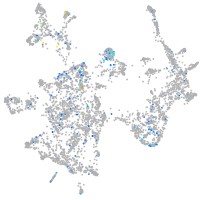
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.227 | tspan36 | -0.082 |
| gpm6aa | 0.220 | gstp1 | -0.072 |
| elavl4 | 0.218 | paics | -0.068 |
| zc4h2 | 0.215 | rab32a | -0.067 |
| stmn1b | 0.210 | rnaseka | -0.066 |
| elavl3 | 0.206 | actb2 | -0.063 |
| ppp1r14ba | 0.204 | akr1b1 | -0.063 |
| tuba1c | 0.197 | si:dkey-151g10.6 | -0.063 |
| mllt11 | 0.194 | prps1a | -0.062 |
| lmo1 | 0.194 | impdh1b | -0.061 |
| tubb5 | 0.193 | slc2a15a | -0.060 |
| nova2 | 0.192 | pts | -0.060 |
| stx1b | 0.192 | zgc:56493 | -0.059 |
| zgc:65894 | 0.190 | syngr1a | -0.058 |
| sncb | 0.190 | rab38 | -0.057 |
| stmn2a | 0.188 | pttg1ipb | -0.057 |
| fez1 | 0.186 | smim29 | -0.056 |
| rtn1a | 0.186 | glulb | -0.056 |
| id4 | 0.181 | tmem130 | -0.055 |
| zgc:153426 | 0.180 | gart | -0.055 |
| rnasekb | 0.179 | pfn1 | -0.055 |
| tmsb | 0.179 | gpr143 | -0.054 |
| si:dkey-12h9.6 | 0.177 | agtrap | -0.054 |
| myt1la | 0.175 | atic | -0.054 |
| gpr85 | 0.174 | mitfa | -0.054 |
| syt1a | 0.173 | gch2 | -0.053 |
| si:ch73-233f7.4 | 0.172 | ppat | -0.053 |
| gpc1a | 0.172 | sdc4 | -0.053 |
| xpr1a | 0.170 | CABZ01032488.1 | -0.053 |
| mab21l1 | 0.170 | ctsba | -0.052 |
| sox4b | 0.170 | C12orf75 | -0.052 |
| ptmaa | 0.169 | pcbd1 | -0.052 |
| nrxn1a | 0.169 | qdpra | -0.052 |
| ptprsb | 0.168 | slc38a5b | -0.052 |
| cd99l2 | 0.167 | mdh1aa | -0.052 |