ankyrin repeat and SOCS box containing 7
ZFIN 
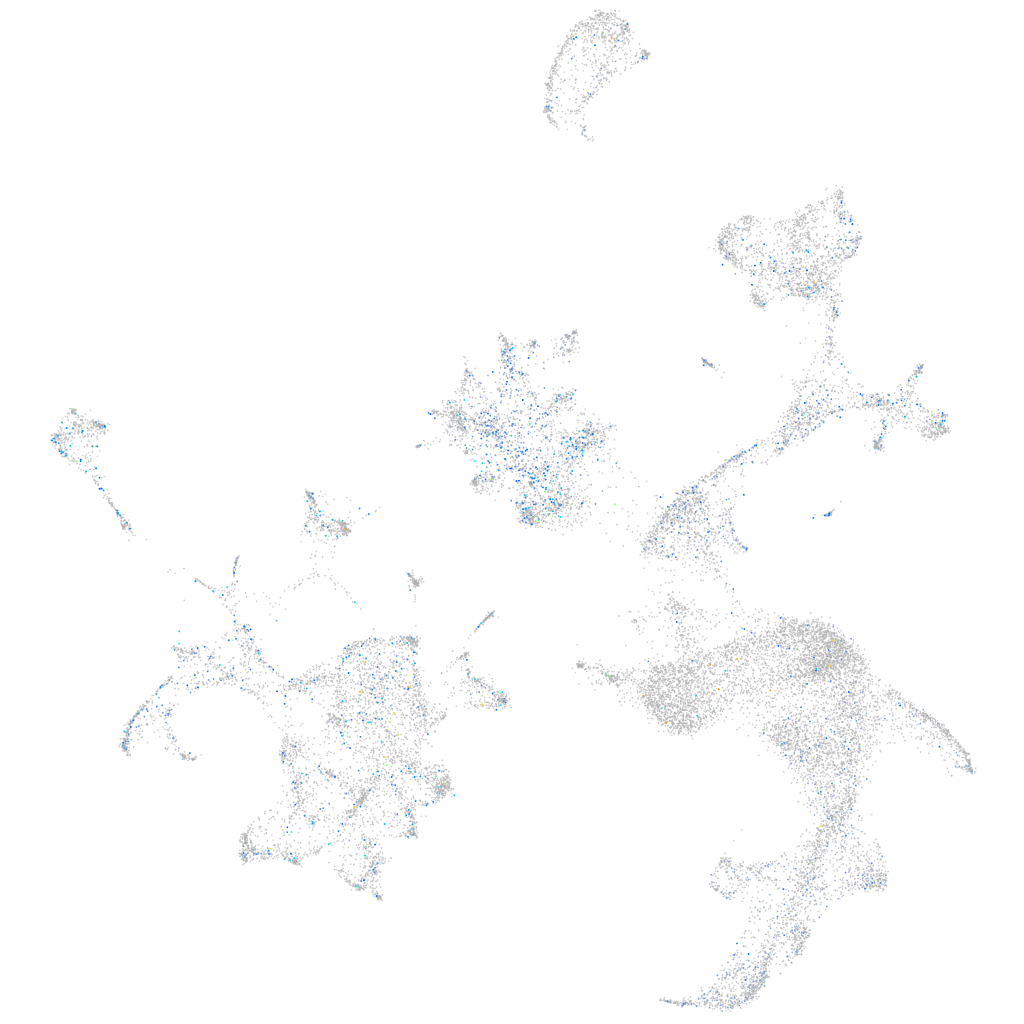
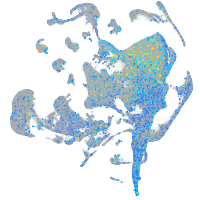
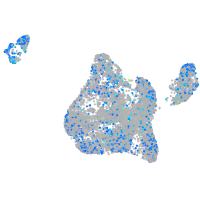
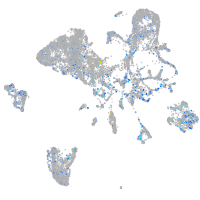
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb1b | 0.127 | cahz | -0.061 |
| hmgn6 | 0.126 | hemgn | -0.059 |
| khdrbs1a | 0.125 | creg1 | -0.058 |
| si:ch211-288g17.3 | 0.125 | hbbe2 | -0.058 |
| ptmab | 0.123 | hbbe1.1 | -0.057 |
| hmgb2b | 0.121 | hbae1.3 | -0.057 |
| tubb2b | 0.121 | hbae3 | -0.055 |
| hnrnpa0b | 0.121 | hbae1.1 | -0.053 |
| hnrnpaba | 0.120 | plac8l1 | -0.053 |
| si:ch73-1a9.3 | 0.116 | hbbe1.3 | -0.053 |
| cirbpa | 0.115 | blvrb | -0.053 |
| tuba1a | 0.114 | nt5c2l1 | -0.052 |
| marcksb | 0.114 | si:ch211-207c6.2 | -0.050 |
| gpm6aa | 0.114 | gpx1a | -0.050 |
| si:ch211-222l21.1 | 0.113 | hbbe1.2 | -0.049 |
| hmgb3a | 0.112 | tspo | -0.048 |
| pfn2l | 0.112 | alas2 | -0.048 |
| chd4a | 0.111 | zgc:163057 | -0.046 |
| hmgn2 | 0.110 | nmt1b | -0.042 |
| hdac1 | 0.110 | tmod4 | -0.042 |
| seta | 0.110 | bloc1s6 | -0.041 |
| syncrip | 0.109 | prdx2 | -0.041 |
| zc4h2 | 0.109 | si:ch211-250g4.3 | -0.040 |
| ilf2 | 0.108 | slc4a1a | -0.038 |
| hp1bp3 | 0.108 | fth1a | -0.038 |
| hmga1a | 0.108 | hbae5 | -0.037 |
| h2afvb | 0.108 | bnip3lb | -0.037 |
| hnrnpa0a | 0.108 | ucp3 | -0.035 |
| hmgn7 | 0.108 | epb41b | -0.034 |
| tubb5 | 0.107 | nt5e | -0.033 |
| sumo3a | 0.107 | selenoj | -0.033 |
| calm2b | 0.106 | faxdc2 | -0.032 |
| si:ch211-133n4.4 | 0.106 | eef2l2 | -0.032 |
| si:ch73-281n10.2 | 0.105 | zgc:56095 | -0.030 |
| elavl3 | 0.105 | gp9 | -0.029 |